Annotate-a-genome: Difference between revisions
Jump to navigation
Jump to search
imported>Weigang |
imported>Weigang |
||
| Line 120: | Line 120: | ||
<syntaxhighlight lang="bash" line start="3" enclose="div"> | <syntaxhighlight lang="bash" line start="3" enclose="div"> | ||
makeblastdb -in accession.pep -parse_seqids # Prepare the new genome DB | makeblastdb -in accession.pep -parse_seqids # Prepare the new genome DB | ||
blastp -query accession.pep -db ref.pep -outfmt 6 -evalue 1e-3 -out accession.fwd # Forward BLAST | blastp -query accession.pep -db ref.pep -outfmt '6 qseqid sseqid pident length qlen evalue' -evalue 1e-3 -out accession.fwd # Forward BLAST # customized outfmt 6 | ||
blastp -query ref.pep -db accession.pep -outfmt 6 -evalue 1e-3 -out accession.rev # Reverse BLAST | blastp -query ref.pep -db accession.pep -outfmt '6 qseqid sseqid pident length qlen evalue' -evalue 1e-3 -out accession.rev # Reverse BLAST | ||
./check-reciprocal.pl <accession.fwd> <accession.rev> > accession.orthlogs 2> accession.not-orthologs # Identify orthologs | ./check-reciprocal.pl <accession.fwd> <accession.rev> > accession.orthlogs 2> accession.not-orthologs # Identify orthologs | ||
</syntaxhighlight> | </syntaxhighlight> | ||
Revision as of 05:13, 9 March 2014
Project Goals
- Annotate and add newly sequenced Borrelia genomes to BorreliaBase
- Build an informatics pipeline for gene prediction, ortholog calls, databasing, and synteny analysis
Claim your assigned genome
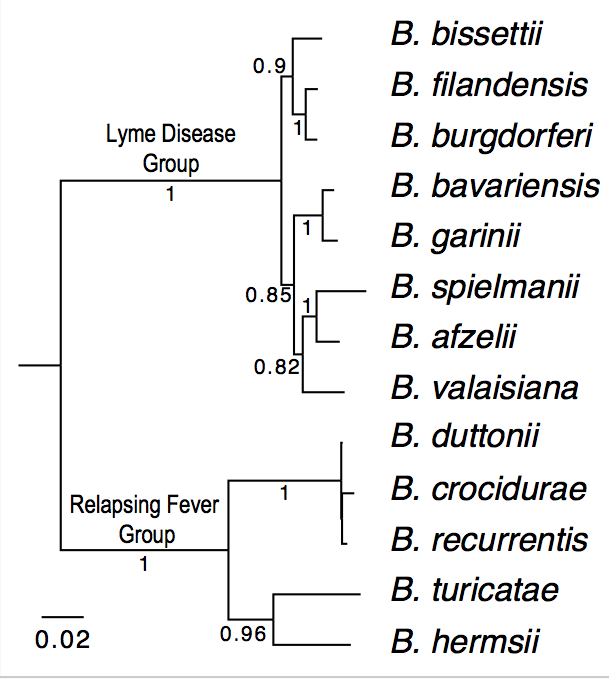
| Genome_id | Strain | Species | Group | Genome Sequences | Notes |
|---|---|---|---|---|---|
| 100 | B31 | B. burgdorferi (reference genome) | Lyme Disease | Reference. Already downloaded as "ref.pep" | |
| 114 | CA382 | B. burgdorferi (California) | Lyme Disease |
| |
| 115 | CA8 | B. burgdorferi (California) | Lyme Disease |
| |
| 304 | BgVir | B. garinii (Russia) | Lyme Disease |
| |
| 305 | NMJW1 | B. garinii (China) | Lyme Disease |
| |
| 402 | HLJ01 | B. afzelii (China) | Lyme Disease |
| |
| 1003 | Ly | B. duttonii (Tanzania) | Relapsing Fever |
|
|
| 1001 | A1 | B. recurrentis (Ethiopia) | Relapsing Fever |
| |
| 1100 | DAH | B. hermsii (Washington State) | Relapsing Fever |
| |
| 1200 | 91E135 | B. turicatae (Texas) | Relapsing Fever |
| |
| 1002 | Achema | B. crocidurae (Mauritania) | Relapsing Fever |
|
|
| 1400 | HR1 | B. parkeri (??) | Relapsing Fever |
| |
| 1300 | LB-2001 | B. miyamotoi (Northeast US) | Relapsing Fever |
| |
| 107 | 94a | B. burgdorferi (Northeast US) | Lyme Disease |
|
Protocol
Dependencies
- BASH (default shell of Linux OS and Apple OS X)
- Perl and BioPerl
- DNATweezer
- NCBI Standalone BLAST+
Part 1. Fetch genome sequences and extract protein sequences
- Note: These scripts are in "../../bio425/annotate-a-genome-pipeline". You may either make a copy to your home directory (recommended) or run directly from that directory by including the path
- Commands:
./fetch-genome.pl <your_assigned_accession> # Expected output: "accession.gb"
./gb2pep.pl <accession.gb> # Expected output: "accession.pep"
Part 2. Predict orthologs with reciprocal BLAST
- Note: Replace the "accession" in the following commands with your assigned accession number.
- Commands:
makeblastdb -in accession.pep -parse_seqids # Prepare the new genome DB
blastp -query accession.pep -db ref.pep -outfmt '6 qseqid sseqid pident length qlen evalue' -evalue 1e-3 -out accession.fwd # Forward BLAST # customized outfmt 6
blastp -query ref.pep -db accession.pep -outfmt '6 qseqid sseqid pident length qlen evalue' -evalue 1e-3 -out accession.rev # Reverse BLAST
./check-reciprocal.pl <accession.fwd> <accession.rev> > accession.orthlogs 2> accession.not-orthologs # Identify orthologs
Part 3. Generate database tables
- Note: Use your assigned genome_id in the table above as the argument for the "-g" option
./gb2table.pl -g <genome_id> -t <accession.gb> # Expected output: "accession.contig.txt"
./gb2table.pl -g <genome_id> -f <accession.gb> # Expected output: "accession.orf.txt"