Bioutils: Difference between revisions
imported>Weigang |
imported>Weigang m (→Demos) |
||
| Line 16: | Line 16: | ||
===Basic Usage=== | ===Basic Usage=== | ||
<syntaxhighlight lang=bash"> | <syntaxhighlight lang=bash"> | ||
bioseq -l foo.fasta # print seq names and lengths | |||
bioseq -s '100, 200' foo.fasta # extract a subsequence | |||
bioseq -r foo.fasta # reverse complement | |||
bioseq -t1 foo.fasta # translate in the +1 frame | |||
bioseq -t3 foo.fasta # translate in +1, +2, and +3 frames | |||
bioseq -t6 foo.fasta # translate in all 6 frames | |||
bioseq -s'100,200' foo.fasta | bioseq -r # subseq and then revcom | |||
bioseq -p'id:seq_1' foo.fasta # pick a sequence by ID | |||
bioseq -p'order:3' # pick the 3rd sequence | |||
bioseq -p're:seq_' foo.fasta # pick a sequence by regular expression | |||
</syntaxhighlight> | </syntaxhighlight> | ||
===Power usage=== | ===Power usage=== | ||
Revision as of 04:16, 29 September 2014
BioPerl-based Sequence Utilities
What is bioutils?
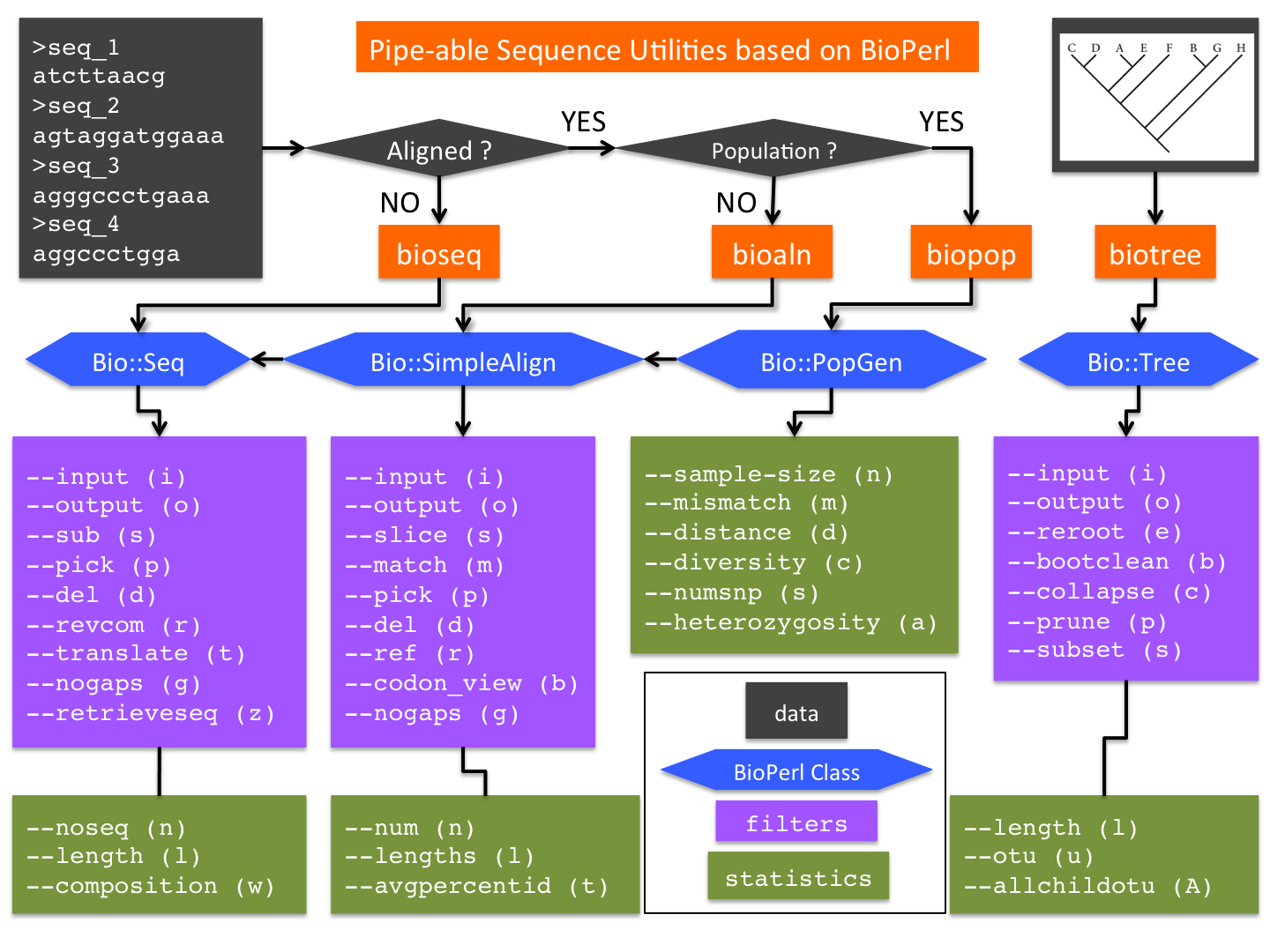
bioutils are a suite of Perl scripts that provide convenient command-line accesses to popular BioPerl methods. Designed as UNIX-like utilities, these tools aim to circumvent the need for composing one-off BioPerl scripts for routine manipulations of sequences, alignments and trees.
The initial release of bioutils consists of four utilities (Fig 1):
- bioseq: a wrapper of BioPerl class Bio::Seq, with additional methods
- bioaln: a wrapper of Bio::SimpleAlign, with additional methods
- biopop: a wrapper of Bio::PopGen, with additional methods
- biotree: a wrapper of Bio::tree, with additional methods
These utilities have been in development since 2002 in the lab of Dr Weigang Qiu at Hunter College of the City University of New York. They are the main code base of the Qiu Lab, which specializes in microbial evolutionary genomics. They proved to be convenient, efficient, and popular among students and researchers. By releasing bioutils as an Open Source tool, we hope to (1) share our experience and (2) invite other developers to join the effort of making BioPerl more accessible.
Demos
Basic Usage
bioseq -l foo.fasta # print seq names and lengths
bioseq -s '100, 200' foo.fasta # extract a subsequence
bioseq -r foo.fasta # reverse complement
bioseq -t1 foo.fasta # translate in the +1 frame
bioseq -t3 foo.fasta # translate in +1, +2, and +3 frames
bioseq -t6 foo.fasta # translate in all 6 frames
bioseq -s'100,200' foo.fasta | bioseq -r # subseq and then revcom
bioseq -p'id:seq_1' foo.fasta # pick a sequence by ID
bioseq -p'order:3' # pick the 3rd sequence
bioseq -p're:seq_' foo.fasta # pick a sequence by regular expressionPower usage
grep -v "Description" ../../bio425/data/ge.dat | wc -l; or: grep -vc "Description" ../../bio425/data/ge.dat
grep "Description" ../../bio425/data/ge.dat | tr '\t' '\n'| grep -v "Desc" | wc -l
grep -Pw "ERBB2|PGR|ESR1" ../../bio425/data/ge.datCreative usage
echo -ne ">lookup\nATG\n" | bioseq -t1 # Lookup a codon productFull documentation
Release notes
Main contributors
- Yozen Hernandez
- Weigang Qiu
- Pedro Pagan
- Levy Vargas