imported>Weigang |
imported>Weigang |
| Line 62: |
Line 62: |
|
| |
|
| ==Full documentation== | | ==Full documentation== |
| <html xmlns="http://www.w3.org/1999/xhtml">
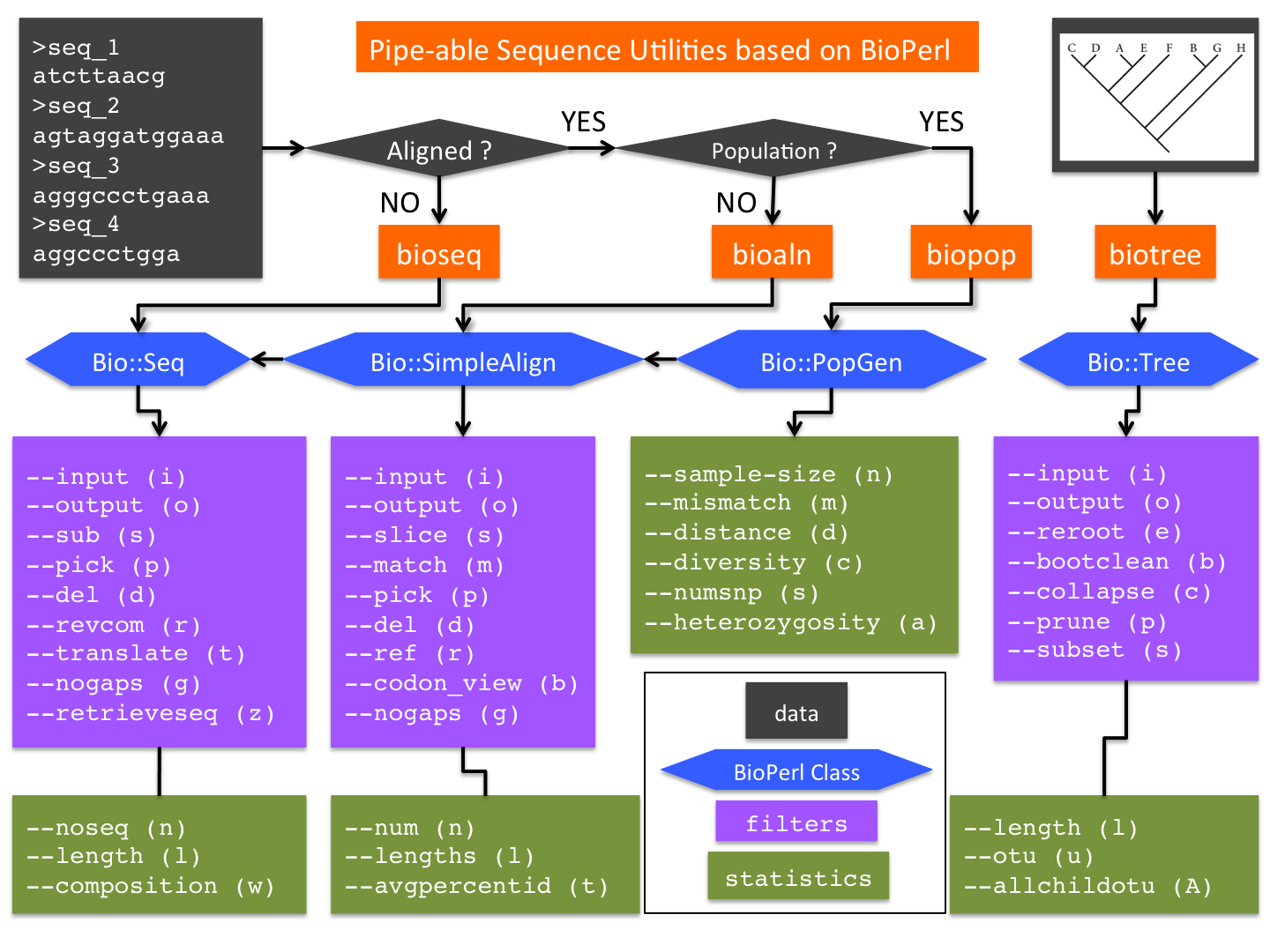
| | * bioseq |
| <head>
| | * bioaln |
| <title>bioseq</title>
| | * biopop |
| <meta http-equiv="content-type" content="text/html; charset=utf-8" />
| | * biotree |
| <link rev="made" href="mailto:root@wallace.hunter.cuny.edu" />
| |
| </head>
| |
| | |
| <body style="background-color: white">
| |
| | |
| | |
| | |
| <ul id="index">
| |
| <li><a href="#NAME">NAME</a></li>
| |
| <li><a href="#SYNOPSIS">SYNOPSIS</a></li>
| |
| <li><a href="#DESCRIPTION">DESCRIPTION</a></li>
| |
| <li><a href="#OPTIONS">OPTIONS</a></li>
| |
| <li><a href="#EXAMPLES">EXAMPLES</a></li>
| |
| <li><a href="#REQUIRES">REQUIRES</a></li>
| |
| <li><a href="#SEE-ALSO">SEE ALSO</a></li>
| |
| <li><a href="#AUTHORS">AUTHORS</a></li>
| |
| <li><a href="#POD-ERRORS">POD ERRORS</a></li>
| |
| </ul>
| |
| | |
| <h1 id="NAME">NAME</h1>
| |
| | |
| <p>bioseq - Fasta sequence editing module based on BioPerl.</p>
| |
| | |
| <h1 id="SYNOPSIS">SYNOPSIS</h1>
| |
| | |
| <p><b>bioseq</b> [options] [sequence file]</p>
| |
| | |
| <h1 id="DESCRIPTION">DESCRIPTION</h1>
| |
| | |
| <p><b>bioseq</b> will read a sequence file and act upon it by doing the following - reformat input (default is fasta) to Genbank or EMBL formats, delete specified sequences, generate overlapping subsequence with a specified window size, generate the reverese complementary sequence, for nucleic acid sequences only, take input list of sequences apart into individual sequence files, extract a specified subset of the sequence, linearize the sequence, remove gaps, find the longest open reading frame (ORF), remove stop codons, give percentage composition of specified amino acids or nuclic acid bases, split the sequences as specified by the user, translate a specific frame of input sequence, or extract a specific gene ID from multiple file sequences. By default, <b>bioseq</b> will assume that both the input and the output are in FASTA format.</p>
| |
| | |
| <h1 id="OPTIONS">OPTIONS</h1>
| |
| | |
| <dl>
| |
| | |
| <dt id="help--h"><b>--help, -h</b></dt>
| |
| <dd>
| |
| | |
| <p>Print a brief help message and exit.</p>
| |
| | |
| <pre><code> Usage: bioseq -h <keyword></code></pre>
| |
| | |
| </dd>
| |
| <dt id="man--m"><b>--man, -m</b></dt>
| |
| <dd>
| |
| | |
| <p>Prints the manual page and exit.</p>
| |
| | |
| <pre><code> Usage: bioseq -m <keyword> </code></pre>
| |
| | |
| </dd>
| |
| <dt id="input--i-format"><b>--input, -i</b> 'format'</dt>
| |
| <dd>
| |
| | |
| <p>Input file format. By default, this is 'fasta'. For Genbank format, use 'genbank'. For EMBL format, use 'embl'.</p>
| |
| | |
| <pre><code> Usage: bioseq -i 'genbank' input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="output--o-format"><b>--output, -o</b> 'format'</dt>
| |
| <dd>
| |
| | |
| <p>Output file format. By default, this is 'fasta'.For Genbank format, use 'genbank'. For EMBL format, use 'embl'.</p>
| |
| | |
| <pre><code> Usage: bioseq -o 'EMBL' input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="noseq--n"><b>--noseq, -n</b></dt>
| |
| <dd>
| |
| | |
| <p>Print number of sequences specified by n.</p>
| |
| | |
| <pre><code> Usage: bioseq -n input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="sub--s-min-max"><b>--sub, -s</b> 'min,max'</dt>
| |
| <dd>
| |
| | |
| <p>Select substring (of 1st sequence),</p>
| |
| | |
| <pre><code> Usage: bioseq -s '<beginning index>, <ending index>' input_file
| |
| | |
| Example: bioseq -s'20,80' input_file (or --sub'20,80' or -s='20,80' or --sub='20,80')</code></pre>
| |
| | |
| </dd>
| |
| <dt id="lengths--l"><b>--lengths, -l</b></dt>
| |
| <dd>
| |
| | |
| <p>Print all sequence lengths.</p>
| |
| | |
| <pre><code> Usage: bioseq -l</code></pre>
| |
| | |
| </dd>
| |
| <dt id="leadgaps--y"><b>--leadgaps, -y</b></dt>
| |
| <dd>
| |
| | |
| <p>Count and return the number of leading gaps in each sequence.</p>
| |
| | |
| <pre><code> Usage: bioseq -y</code></pre>
| |
| | |
| </dd>
| |
| <dt id="pick--p-tag:value"><b>--pick, -p</b> 'tag:value'</dt>
| |
| <dd>
| |
| | |
| <p>Select a single sequence or a comma-separated list of sequences, e.g, --pick 'id:foo' by id --pick 'order:2' by order --pick 're:REGEX' using a regular expression (only one regex is expected)</p>
| |
| | |
| <p>Specifically for a list of sequences, --pick 'id:foo,bar' list by id --pick 'order:2,3' list by order --pick 'order:2-10' list by range</p>
| |
| | |
| <pre><code> Usage: bioseq -p 'id:<number>' input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="del--d-tag:value"><b>--del, -d</b> 'tag:value'</dt>
| |
| <dd>
| |
| | |
| <pre><code> Delete a sequence or a comma-separated list of sequences, eg,
| |
| --del 'id:foo' by id
| |
| --del 'order:2' by order
| |
| --del 'length:n' by min length, where 'n' is length
| |
| --del 'ambig:x' by min % ambiguous base/aa, where 'x' is the %
| |
| --del 'id:foo,bar' list by id
| |
| --del 're:REGEX' using a regular expression (only one regex is expected)
| |
| | |
| Usage: bioseq --del 'id:<number>' input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="revcom--r"><b>--revcom, -r</b></dt>
| |
| <dd>
| |
| | |
| <p>Reverse complement</p>
| |
| | |
| <pre><code> Usage: bioseq -r input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="slidingwindow--k-window_size"><b>--slidingwindow, -k</b> 'window_size'</dt>
| |
| <dd>
| |
| | |
| <p>Generate overlapping subsequence with a specified window size (default 1)</p>
| |
| | |
| <pre><code> Usage: bioseq -k '<index of subsequence>' input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="translate--t-1-3-6"><b>--translate, -t</b> [1|3|6]</dt>
| |
| <dd>
| |
| | |
| <p>Translate in 1, 3, or 6 frames. eg, -t1, -t3, or -t6.</p>
| |
| | |
| <pre><code> Usage: bioseq -t3 input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="shred--c"><b>--shred, -c</b></dt>
| |
| <dd>
| |
| | |
| <p>Shred into individual sequences</p>
| |
| | |
| <pre><code> Usage: bioseq -c input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="linearize--L"><b>--linearize, -L</b></dt>
| |
| <dd>
| |
| | |
| <p>Linearize FASTA.</p>
| |
| | |
| <pre><code> Usage: bioseq -L fasta_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="extract--e"><b>--extract, -e</b></dt>
| |
| <dd>
| |
| | |
| <p>Extract in-frame sequences.</p>
| |
| | |
| <pre><code> Usage: bioseq -e input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="nogaps--g"><b>--nogaps, -g</b></dt>
| |
| <dd>
| |
| | |
| <p>Remove gaps</p>
| |
| | |
| <pre><code> Usage: bioseq -g input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="longest-orf--C"><b>--longest-orf, -C</b></dt>
| |
| <dd>
| |
| | |
| <p>Output the frame that gives the longest ORF.</p>
| |
| | |
| <pre><code> Usage: bioseq -C input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="removestop--x"><b>--removestop, -x</b></dt>
| |
| <dd>
| |
| | |
| <p>Remove stop codons (for e.g., PAML input)</p>
| |
| | |
| <pre><code> Usage: bioseq -x input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="anonymize--a-n"><b>--anonymize, -a</b> 'n'</dt>
| |
| <dd>
| |
| | |
| <p>Replace sequence IDs with serial IDs 'n' characters long, including a leading 'S' (e.g., -a'5' gives S0001). Produces a sed script file with a '.sed' suffix that may be used with sed's '-f' argument. If the filename is '-', the sed file is named STDOUT.sed instead. The sed filename is specified on STDERR.</p>
| |
| | |
| <pre><code> Usage: bioseq -a <number> input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="prefix-PREFIX"><b>--prefix</b> 'PREFIX'</dt>
| |
| <dd>
| |
| | |
| <p>Used in conjunction with --anonymize. This lets you specify a custom prefix for the anonymized sequence IDs given when using the --anonymize option. If this is given, then the whole prefix will count toward the total ID length. For example: suppose the prefix chosen is SEQ, and that for --anonymize you supplied 5. Then the maximum id length is 5, so there is room for only two more digits. e.g., SEQ01 atg... SEQ02 atg... SEQ03 atc...</p>
| |
| | |
| <p>If there are enough sequences that the length of the prefix plus the length of the digit portion exceeds the length given to --anonymize, a warning will be given: aln-manipulations.pl -a 4 --prefix=SEQ # output SEQ1 atg... ... SEQ10 atc...</p>
| |
| | |
| <pre><code> # more output
| |
| WARNING: Anonymized ID length exceeded requested length: try a different length or prefix.</code></pre>
| |
| | |
| </dd>
| |
| <dt id="composition--w"><b>--composition, -w</b></dt>
| |
| <dd>
| |
| | |
| <p>Base or AA composition.</p>
| |
| | |
| <pre><code> Usage: bioseq -w input_file</code></pre>
| |
| | |
| </dd>
| |
| <dt id="split--S-split_at"><b>--split, -S</b> 'split_at'</dt>
| |
| <dd>
| |
| | |
| <p>Split the input sequence file into several smaller files with 'split_at' sequences per file. For instance, to split a file into several smaller files with 100000 sequences each, you would run:</p>
| |
| | |
| <pre><code> bioseq -S 100000 seq.fasta</code></pre>
| |
| | |
| <p>The output files will be named with the label "split_N" according to the input file (or with the STDIN prefix if the file is read via standard in), where N denotes the "part" or "split" number.</p>
| |
| | |
| </dd>
| |
| <dt id="retrieveseq--z-sequence-retriever-using-GenBank-accession"><b>--retrieveseq, -z</b> 'sequence retriever using GenBank accession'</dt>
| |
| <dd>
| |
| | |
| <pre><code> Usage: bioseq -z [Accession]
| |
| Retrieves the sequence from GenBank using the provided GenBank accession. Prints out text in a fasta file.
| |
| | |
| EXAMPLE:
| |
| | |
| bioseq -z X83553
| |
| | |
| OUTPUT:
| |
| | |
| >X83553 B.garinii (PHei strain) opsC gene.
| |
| ATGAAAAAGAATACATTAAGTGCGATATTAATGACTTTATTTTTATTTATATCTTGTAAT
| |
| AATTCAGGTGGGGATACTGCATCTACTAATCCTGATGAGTCTGCGAAAGGACCTAATCTT
| |
| ATAGAAATAAGCAAAAAAATTACAGATTCTAATGCATTTGTACTGGCTGTGAAAGAAGTT
| |
| GAGGCTTTGATCTCATCTATAGATGAACTTGCTAATAAAGCTATTGGTAAAAAAATAAAT
| |
| CAAAATGGTTTAGATGCTGATGCTAATCACAACGGATCATTGTTAGCAGGAGCCCATGCA
| |
| ATATCAACTCTAATAAAACAAAAAACAGATGGATTGAAAGATCTAGAAGGGTTAAGTAAA
| |
| GAAATTGCAAAGGTGAAGGAATGTTCCGATAAATTTACTAAAAAGCTAACAGATAGTCAT
| |
| GCACAGCTTGGAGCAGTTGGTGGTGCTATTAATGATGATCGTGCAAAAGAAGCTATTTTA
| |
| AAAACACATGGGACTAACGATAAGGGTGCTAAAGAACTTAAAGAGTTATCTGAATCAGTA
| |
| GAAAGCTTGGCAAAAGCAGCTCAAGCAGCATTAGCTAATTCAGTTAAAGAGCTTACAAGT
| |
| CCTGTTGTGGCAGAAAGTCCAAAAAAACCTTAA</code></pre>
| |
| | |
| </dd>
| |
| <dt id="dotplot--D-draw_dotplot"><b>--dotplot, -D</b> 'draw_dotplot'</dt>
| |
| <dd>
| |
| | |
| <p>Extract two sequences from input file and generate a dotplot.</p>
| |
| | |
| <pre><code> Usage: bioseq -D 'id1,id2,window_size,slider' fasta_file
| |
| </code></pre>
| |
| | |
| <p>Id1 and Id2 are extracted with their corresponding sequences. Be sure to use the entire sequence identifer, as this is a whole string match. Window_size corresponds to the number of character you would like to compare (Default window is 10). Slider is the number of windows to compare (Default slider is 10). The sequence corresponding to ID1 will appear on the X axis (row) and ID2 on the Y axis (column). This method will work on both DNA and Amino Acids.</p>
| |
| | |
| <pre><code> Example:
| |
| Sample Input:
| |
| >id1
| |
| ATACGA
| |
| >id2
| |
| ATACGA
| |
| | |
| Command: bioseq -D 'id1,id2,3'
| |
| Output:
| |
| | |
| A T A C G A
| |
| A *
| |
| T *
| |
| A *
| |
| C
| |
| G
| |
| A
| |
| A
| |
| C
| |
| A
| |
| A
| |
| T
| |
| G</code></pre>
| |
| | |
| <p>==item <b>--rename, -R</b> 'rename_id'</p>
| |
| | |
| <pre><code> Usage: bioseq -R [file_with_new_names] file_to_be_changed.fasta
| |
| | |
| Ex: bioseq -D list.txt test-bioseq.nuc
| |
| | |
| Input:
| |
| | |
| list.txt:
| |
| | |
| VS116:7:310:IGS:11 VS116
| |
| B31:1:100:IGS:11 B31
| |
| | |
| *Left column is the pattern to be replaced by the right column
| |
| | |
| file_to_be_changed.fasta:
| |
| | |
| >VS116:7:310:IGS:11
| |
| AATTTCAAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTGAAATTTTTCTAT
| |
| TGGATAAATTCTATACAAGAAGGTAAATA
| |
| >B31:1:100:IGS:11
| |
| AATTTTTAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTTAAAACTTTTCTA
| |
| TTGGATAGATTTTATACAAAGAAGGTAATA
| |
| | |
| Output:
| |
| >VS116
| |
| AATTTCAAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTGAAATTTTTCTAT
| |
| TGGATAAATTCTATACAAGAAGGTAAATA
| |
| >B31
| |
| AATTTTTAAAATATAATATAAAAACAGCTAATCCAATAGAAAAATTTTAAAACTTTTCTA
| |
| TTGGATAGATTTTATACAAAGAAGGTAATA</code></pre>
| |
| | |
| </dd>
| |
| </dl>
| |
| | |
| <h1 id="EXAMPLES">EXAMPLES</h1>
| |
| | |
| <p>Section under construction...</p>
| |
| | |
| <h1 id="REQUIRES">REQUIRES</h1>
| |
| | |
| <p>Perl 5.010, BioPerl</p>
| |
| | |
| <h1 id="SEE-ALSO">SEE ALSO</h1>
| |
| | |
| <pre><code> perl(1)</code></pre>
| |
| | |
| <h1 id="AUTHORS">AUTHORS</h1>
| |
| | |
| <pre><code> Weigang Qiu at genectr.hunter.cuny.edu
| |
| Yözen Hernández yzhernand at gmail dot com
| |
| Levy Vargas levy dot vargas at gmail dot com</code></pre>
| |
| | |
| <h1 id="POD-ERRORS">POD ERRORS</h1>
| |
| | |
| <p>Hey! <b>The above document had some coding errors, which are explained below:</b></p>
| |
| | |
| <dl>
| |
| | |
| <dt id="Around-line-382">Around line 382:</dt>
| |
| <dd>
| |
| | |
| <p>Non-ASCII character seen before =encoding in 'Yözen'. Assuming UTF-8</p>
| |
| | |
| </dd>
| |
| </dl>
| |
| | |
| | |
| </body>
| |
| | |
| </html>
| |
| ---- | | ---- |
|
| |
|