QuBi/modules/biol203-Lab4: Difference between revisions
imported>Weigang |
imported>Weigang |
||
| Line 73: | Line 73: | ||
## Align the first 5 bases of all introns. Which bases are conserved near intron start ("donor site")? | ## Align the first 5 bases of all introns. Which bases are conserved near intron start ("donor site")? | ||
## Align the last 5 bases of all introns. Which bases are conserved near intron end ("acceptor site")? | ## Align the last 5 bases of all introns. Which bases are conserved near intron end ("acceptor site")? | ||
## Follow examples and instructions on [http://weblogo.berkeley.edu/ WebLogo] and make a | ## Follow examples and instructions on [http://weblogo.berkeley.edu/ WebLogo] and make a sequence logo for the acceptor site and another sequence logo for the donor site. | ||
<center> | <center> | ||
Table 1. ''mdm2'' Exons | Table 1. ''mdm2'' Exons | ||
Revision as of 13:53, 11 February 2015
Lab 4. Bioinformatics Exercises: BLAST & Gene Structure
Expected Learning Outcomes
- Be able to perform NCBI BLAST search for homologous sequences in GenBank.
- Be able to identify individual gene elements based on an NCBI GenBank file.
- Be able to identify alternative splice forms a single gene using NCBI web tools
Lab Report Grading Policy
- Introduction (5 pts). Define these terms: bioinformatics, homology, BLAST, e-value, alternative splicing (Your statements are not to be copied from the Lab Manual.)
- Materials and Methods (5 pts). List and describe steps of a BLAST search & steps of identifying alternative splicing variants with BLAST.
- Results (20 pts). Copy your results in a document during the exercises, and hand in an organized copy. Include answers to all queries and questions.
- Discussion and Conclusion (15 pts). Answer the five discussion questions. Summary/Conclusion: a sentence or two will suffice.
- References (5 pt). Credit is given for pertinent references obtained from sources other than the Lab Manual.
(Total: 50 pts)
Introduction
Research in molecular genetics requires effective use of online bioinformatic tools to analyze and understand the genetic materials being worked with. The following exercises will expose you to real-world scenarios and introduce you to the methods and tools you can use to solve these problems.
In biology, homology is defined as a common or shared evolutionary origin. Therefore, homologous sequences are sequences diverged from a common ancestor. Note that the word "homology" is different from "similarity": homologous structures or sequences may not be similar (e.g., forearms in mammals and birds) and, conversely, similar structures or sequences may not be homologous (e.g., wings in birds and bats).
BLAST is a computer algorithm allowing for efficient search of similar sequences in a large database. While BLAST performs a similar function to Google search, you should not use Google to look for similar sequences in a human or other genome. When sequences are similar with a sufficient statistical significance (measured by e-value, see below), we consider these sequences homologous to each other.
Exercise 1. Homology searching using BLAST
- Go to the NCBI-BLAST website at NCBI/BLAST Home Page
- What is BLAST? Read and copy the expanded answer by clicking on "more"
- Since BLAST finds matches between biological sciences, it needs a "query" sequence as input as well as a "database" to search against. To find matches of a sequence in human genome, what would be your "query" sequence and what would be your "database"?
- Start BLASTing against the mouse genome by clicking "Mouse" under "BLAST Assembled RefSeq Genomes"
- Copy and paste the following sequence into the "Enter Query Sequence" box:
CTAGATGCATTTACGAAGGAGACAGAAAACGTCTTTCGGCAATAGCTCTCAAATGCAAAACGACGTCGG CGAGCTGTCCCTTACCTGGAGGCCCGCAGGAGAAGCGCGGTGATCCGAGAGGGTCCCCCAGGGGTGTCCG GTCGGTCTCCCGCTCGCCCAGCAGACGGCTGCGGAAACGGGGCAGCGTTTAAATAACCCCAGCTGGAGAC ATGTCAGGACTTAGCTCCTCCGACAGCCGACGCCGGACGTGTCCCAACTTGACCAGCCCCACAGGAAGAG CTGAGTCAACTCGGCCCAGCCCAGTCCCACCCGTCCCGGAAGCCGCATCCCGGCGAGTCCGGGACCAGGC ACCTGTCACCTCCTGGACCCCAGCAACGAGCCCAGCGCGACCCCGGAGCGGGCCCGAATTCT
- Scroll down to the bottom of the page and click "BLAST"
- Wait for 10-30 seconds for the results to return (be patient). Once the result page is loaded, locate and copy/write down
the following information for the first hit:
- Species and strain
- Chromosome
- Length of your query sequence
- Sequence identity, number of matched bases, and number of gaps between the matched sequences
- Click the link for "5' side" (next to Features) will bring you a standard GenBank file of this gene. Locate and copy the
following structural information about this gene:
- Gene accession (ID number)
- Total length of the gene
- Number of introns
- Which is the non-template (mRNA analog) strand: the above sequence itself or its reverse complement? [Hint: note the word complement in mRNA and cDNA lines)
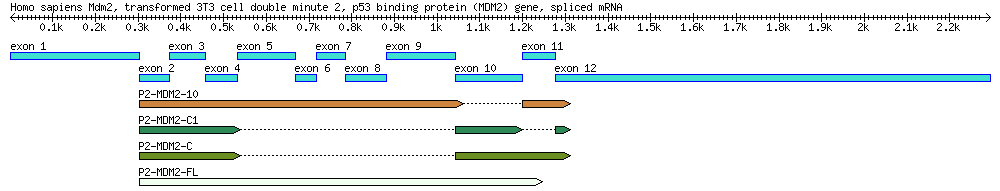
Exercise 2. Explore the structure of human mdm2 gene
- Search GenBank using the accession AF527840. Read the GenBank file and find out from the feature table how many introns and exons this sequence has according to the "mRNA" and "CDS" features.
- Click on "mRNA" and notice that exon sequences are now highlighted
- Fill in Table 1 for each EXON you could identify:
- Fill in Table 2 for each INTRON you could identify:
- Click on "CDS" and notice that coding sequences are now highlighted
- Fill in Table 3 for each coding sequence you could identify:
- DRAW a diagram of this gene based on the above exon and intron coordinates.
- Label the top of the diagram with basic information, such as the gene's name and species information.
- Label coordinates for introns, exons, 3'/5' UTRs, start-codon, and stop-codon coordinates.
- Draw the diagram mostly to scale. It does NOT have to be perfect, but make a reasonable effort. Put a scale bar and length markers on your drawing.
- Answer the following questions:
- What is the total length of exons, introns, and coding sequences of this gene?
- Are all exon sequences code for proteins? Which exons are non-coding in mdm2?
- Align the first 5 bases of all introns. Which bases are conserved near intron start ("donor site")?
- Align the last 5 bases of all introns. Which bases are conserved near intron end ("acceptor site")?
- Follow examples and instructions on WebLogo and make a sequence logo for the acceptor site and another sequence logo for the donor site.
Table 1. mdm2 Exons
| Exon # | Start Position | End Position | Length |
|---|---|---|---|
| #1 | 1971 | 2271 | 301 |
| #2 | ? | ? | ? |
Table 2. mdm2 Introns
| Intro Number | Start Position | End Position | Length | First 5 bases | Last 5 bases | Phase* |
|---|---|---|---|---|---|---|
| #1 | 2272 | 2987 | 616 | GTACT | TGTAG | ? |
| #2 | ? | ? | ? | ? | ? | ? |
- Introns have phases. Phase 0 introns sit between 2 codons, phase 1 intron sit between the 1st codon position and the 2nd codon position, and phase 3 introns sit between the 2nd and 3rd codon position. How would you find out the phase of an intron? [Hint, use Table 3 CDS positions below].
Table 3. mdm2 Coding Sequences (CDS)
| CDS # | Start Position | End Position | Length |
|---|---|---|---|
| #1 | 2992 | 3072 | 81 |
| #2 | ? | ? | ? |
Exercise 3. Alternative splicing: Use BLAST to determine which exons are present in an mRNA
You will use the following table for your exercise:
| Genbank Accession # | cDNA Clone | Description | Cell Line | Length (bp) |
|---|---|---|---|---|
| AF527840 | Genomic DNA | 34,088 | ||
| EU076746 | P2-MDM2-C1 | cDNA | MANCA | 427 |
| EU076747 | P2-MDM2-10 | cDNA | ML-1 | 842 |
| EU076748 | P2-MDM2-C | cDNA | A876 | 505 |
| EU076749 | P2-MDM2-FL | cDNA | SJSA-1 | 845 |
Blast one of the mRNA sequences (EU076746, EU076747, EU076748, EU076749) against the main sequence (AF527840) and use the results to answer the following questions. Suggested procedures:
- Go to the NCBI BLAST website
- Click the link “Align two (or more) sequences using BLAST (bl2seq)” under “Specialized BLAST” (near the page bottom)
- In the “Sequence 1” text box, type in "EU076748" (or other cDNA accession in the table). In the “Sequence 2” text box, type “AF527840” (the accession for the genomics).
- Click “Align”. You should get a “Blast Result” output page.
- Fill in the following table based on BLAST-identified coordinates:
Table 4. A splice variant of mdm2 (Your choice of mRNA accession:________)
| Match # | Query start | Query end | Subject start | Subject end | Exon # (consult Table 1) |
|---|---|---|---|---|---|
| ? | ? | ? | ? | ? | ? |
Discussion Questions
- Explain the following BLAST terms: “Expect” (e-value) (read this FAQ), “Identities”, “Gap”, “Strand”.
- Which is a statistically more significant match by BLAST, a match with an e-value=1e-5 or a match with an e-value of 1?
- If you want your match to be biologically relevant (and not random, chance matches), should you use the default e-value cutoff of 10?
- List and describe individual elements of a typical human gene based on mdm2.
- What is the "GT-AG" rule? Explain how to read the sequence logos. Explain the significance of sequence conservation at exon-intron junctions.
- Describe biological significance of alternative splicing, using mdm2 gene as an example.
Reference
- Arva NC, Talbott KE, Okoro DR, Brekman A, Qiu WG, Bargonetti J. 2008. Disruption of the p53-Mdm2 complex by Nutlin-3 reveals different cancer cell phenotypes. Ethnicity and Disease. 18(S2):1-8. PubMed Abstract