Summer 2021: Difference between revisions
Jump to navigation
Jump to search
imported>Weigang |
imported>Weigang m (→August 2021) |
||
| (49 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Group schedule== | ==Group meeting/Field trip schedule== | ||
[[File:Beaver-lake.jpg|thumbnail|Quabbin Reservoir & Beaver Lake, MA]] | |||
===August 2021=== | |||
* Wrapping up: Bb transcriptomes, [https://www.rdocumentation.org/packages/plotly/versions/4.9.4.1/topics/ggplotly make interactive plots using ggplotly]. Interactive volcano plots: | |||
** [http://diverge.hunter.cuny.edu/~weigang/Drecktraph_starvation.html Starvation response: differentially expressed genes]. Work credit (data collection, R analysis, and web-interactive visualization): Hannah Ford. Data source: [https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1005160 Drecktah et al (2015). PLoS Pathogen] | |||
** [http://diverge.hunter.cuny.edu/~weigang/drecktrah_interactive.html Borrelia starvation response: differentially expressed sRNA]. Work credit: Jackie Yee. Data source: [https://www.frontiersin.org/articles/10.3389/fcimb.2018.00231/full Drecktrah et al (2018).] | |||
** [http://diverge.hunter.cuny.edu/~weigang/caskey_interactive.html Response to Doxycycline treatment]. Work credit: Jacki Yee. Data source: [https://www.frontiersin.org/articles/10.3389/fmicb.2019.00690/full Caskey et al (2019)] | |||
* Monte Carlo Club Season IV. [[Monte_Carlo_Club | Machine learning with Scikit-Learn]] | |||
* First publication on computational vaccine design (with a blog post): [https://go.nature.com/3xAxOYh Jenner's Dilemma]. | |||
===July 2021=== | |||
* Belfer housekeeping duties: During the month of July, members of Qiu lab (including the PI) will be responsible for the following: | |||
# Performing a careful sweep of the common seating, eating, and food preparation areas each Monday, Wednesday, and Friday. The purpose of this sweep is to do some light organization/cleaning and to identify persistent cleanliness problems that should then be reported (via the lab’s PI) to myself and Elizabeth Cohn (ec2692@hunter.cuny.edu), and the collective PIs of the floor. | |||
# Throwing away any personal food (and personal food containers) left in the refrigerators beyond 5 pm on each Friday. | |||
# Keeping general tabs on the bathrooms of the floor to ensure cleanliness. Persistent cleanliness problems that are unreasonable for the WCMC custodial staff should be addressed by the members of the floor. | |||
* July 2, Friday. Field trip | |||
** location: Rockefeller State Park | |||
** Participants: Saymon, Lia, Joy, Weigang | |||
* Lab meeting: Weekly on Tuesday 11-1 | |||
===June 2021=== | |||
* June 3, 2021 (Thursday). Summer research kickoff | * June 3, 2021 (Thursday). Summer research kickoff | ||
* June 8, 2021 (Tuesday). NLP models of protein structure (Eamen, Roman, Edgar) | * June 8, 2021 (Tuesday). 11-2 | ||
* June 10, 2021 (Thursday). | ** Algorithm development (Brian) | ||
** NLP models of protein structure (Eamen, Roman, Edgar) | |||
** Bb transcriptomics (Niemah & Jackie) | |||
** HIV compartmentalization (Lily) | |||
* June 10, 2021 (Thursday). No meeting. Field day | |||
** Location: Hackscher State Park, Long Island | |||
** Participants: Desiree, Lily, Lia, John, Weigang | |||
** Outcome: ~60 nymph ticks | |||
* June 15, 2021 (Tuesday). 11-2. Lab meeting | |||
* June 17, 2021 (Thursday). No meeting. Field day | |||
** Location: Quabbin Reservoir, MA | |||
** Participant: Saymon, Brian, Lia, Weigang | |||
** Outcome: ~130 nymph ticks | |||
* June 22, 2021 (Tuesday). 11-1 | |||
** Niemah & Jackie will work on collecting transcriptome data into Excel sheets | |||
** Lily presented latest trees on HIV compartmentalization | |||
** Roman will work on env gene embedding using Facebook pretrained ESM model | |||
* June 24, 2021 (Thursday). Lab meeting 11-1 | |||
* June 30, 2021 (Wed). Lab meeting 11-1 | |||
==Project 1. Borrelia genomics== | ==Project 1. Borrelia genomics== | ||
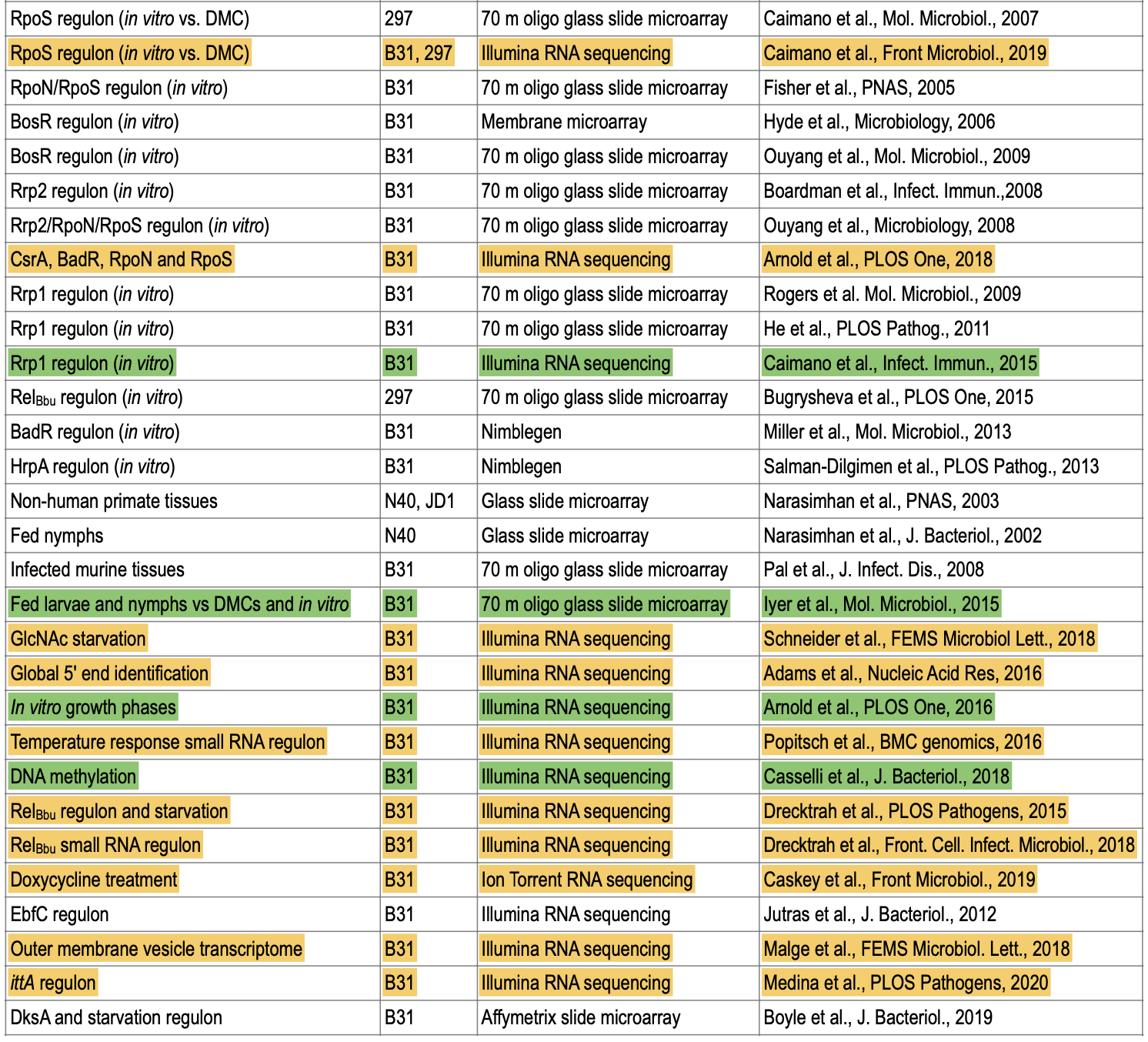
[[File:Transcriptome-table.png|thumbnail]] | |||
* Participants: Niemah, Jackie | * Participants: Niemah, Jackie | ||
* Questions & Goals: | * Questions & Goals: | ||
** Upgrade database, genome pipeline, and website (Lia) | ** Upgrade database, genome pipeline, and website (Lia) | ||
** Phylogeography & evolutionary maintenance of divided genome (Saymon) | ** Phylogeography & evolutionary maintenance of divided genome (Saymon) | ||
** vls evolution (with simulation) & development of immunoflorescence microsopy methods(Lily) | ** vls evolution (with simulation) & development of immunoflorescence microsopy methods(Lily). [https://www.caister.com/openaccess/pdf/9781913652616-17.pdf Live imaging.] | ||
* Reading list | * Reading list | ||
** Latest review book [https://www.caister.com/lyme Lyme Disease and Relapsing Fever Spirochetes: Genomics, Molecular Biology, Host Interactions and Disease Pathogenesis]. [https://www.caister.com/openaccess/pdf/9781913652616-05.pdf The chapter on gene regulation and transcriptomics] (notice Fig 1, Fig 2, and Table 1) | ** Latest review book [https://www.caister.com/lyme Lyme Disease and Relapsing Fever Spirochetes: Genomics, Molecular Biology, Host Interactions and Disease Pathogenesis]. [https://www.caister.com/openaccess/pdf/9781913652616-05.pdf The chapter on gene regulation and transcriptomics] (notice Fig 1, Fig 2, and Table 1) | ||
** Schward et al (2021). [https://pubmed.ncbi.nlm.nih.gov/33328355/ Multipartite Genome of Lyme Disease Borrelia: Structure, Variation and Prophages ] | ** Schward et al (2021). [https://pubmed.ncbi.nlm.nih.gov/33328355/ Multipartite Genome of Lyme Disease Borrelia: Structure, Variation and Prophages ] | ||
** Stevenson & Seshu (2018). [https://pubmed.ncbi.nlm.nih.gov/29064060/ Regulation of Gene and Protein Expression in the Lyme Disease Spirochete ] | ** Stevenson & Seshu (2018). [https://pubmed.ncbi.nlm.nih.gov/29064060/ Regulation of Gene and Protein Expression in the Lyme Disease Spirochete ] | ||
** The bowtie model of gene regulation: [https://pubmed.ncbi.nlm.nih.gov/28767643/ Yan et al (2017). PLoS Comp Biol.] | |||
==Project 2. Design algorithms for vaccines== | ==Project 2. Design algorithms for vaccines== | ||
| Line 19: | Line 59: | ||
* Questions & Goals: | * Questions & Goals: | ||
** Generalized algorithms for antigen with arbitrary tree shape | ** Generalized algorithms for antigen with arbitrary tree shape | ||
*** [https://scikit-learn.org/stable/modules/generated/sklearn.cluster.KMeans.html Scikit-learn K-means] | |||
*** Data set 1. Neutral evolution (with exponentially distributed branch lengths). Binary strings (L=100 bits) evolved from a coalescent tree of 20 leaves. Simulated with <code>rcoal(20); rTraitDisc; simSeq()</code>. [[Monte_Carlo_Club#Summer_Project_1._Systems_evolution_of_biofilm.2Fswarming_pathway_.28with_Dr_Joao_Xavier_of_MSKCC.29|code from previous work]] | *** Data set 1. Neutral evolution (with exponentially distributed branch lengths). Binary strings (L=100 bits) evolved from a coalescent tree of 20 leaves. Simulated with <code>rcoal(20); rTraitDisc; simSeq()</code>. [[Monte_Carlo_Club#Summer_Project_1._Systems_evolution_of_biofilm.2Fswarming_pathway_.28with_Dr_Joao_Xavier_of_MSKCC.29|code from previous work]] | ||
*** Data set 2. Two major clades. HA sequences from fluB | *** Data set 2. Two major clades. HA sequences from fluB | ||
| Line 27: | Line 68: | ||
** Naive Bayes models to integrate immunogenicity data | ** Naive Bayes models to integrate immunogenicity data | ||
** Natural language models to improve structural stability (see Project 4 below) | ** Natural language models to improve structural stability (see Project 4 below) | ||
** Improve protein solubility with Solubis | |||
** Estimating Ag-Ab binding parameters with hierarchical Bayesian modeling: | |||
*** [https://www.r-bloggers.com/2016/11/hierarchical-models-with-rstan-part-1/ a tutorial of R-Stan]; [https://mc-stan.org/docs/2_27/stan-users-guide/index.html Stan manual] | |||
*** [https://r-nimble.org/html_manual/cha-lightning-intro.html with R-Nimble] | |||
*** [https://medium.com/@ODSC/hierarchical-bayesian-models-in-r-9a18e6acdf2b a short tutorial with JAGS] | |||
*** [https://onlinelibrary.wiley.com/doi/pdf/10.1002/0470863692.app1 A workout commercial banking example] | |||
*** [https://www.mrc-bsu.cam.ac.uk/software/bugs/ BUGS; Win-BUGS, and Open-Bugs] | |||
* Reading list | * Reading list | ||
** Greet De Baets, Joost Van Durme, Rob van der Kant, Joost Schymkowitz, Frederic Rousseau, Solubis: optimize your protein, Bioinformatics, Volume 31, Issue 15, 1 August 2015, Pages 2580–2582, https://doi.org/10.1093/bioinformatics/btv162 | |||
** [https://www.nature.com/articles/s41598-019-46740-5 Degoot et al (2019). Predicting Antigenicity of Influenza A Viruses Using biophysical ideas] | |||
** [https://www.biorxiv.org/content/10.1101/2020.12.16.423180v1 Di et al (2021). Maximum antigen divergence in Lyme bacterial population] | ** [https://www.biorxiv.org/content/10.1101/2020.12.16.423180v1 Di et al (2021). Maximum antigen divergence in Lyme bacterial population] | ||
** Golovanov et al (2004). [https://pubs.acs.org/doi/10.1021/ja049297h A Simple Method for Improving Protein Solubility and Long-Term Stability. J. Am. Che. Soc.] | |||
==Project 3. HIV compartmentalized evolution== | ==Project 3. HIV compartmentalized evolution== | ||
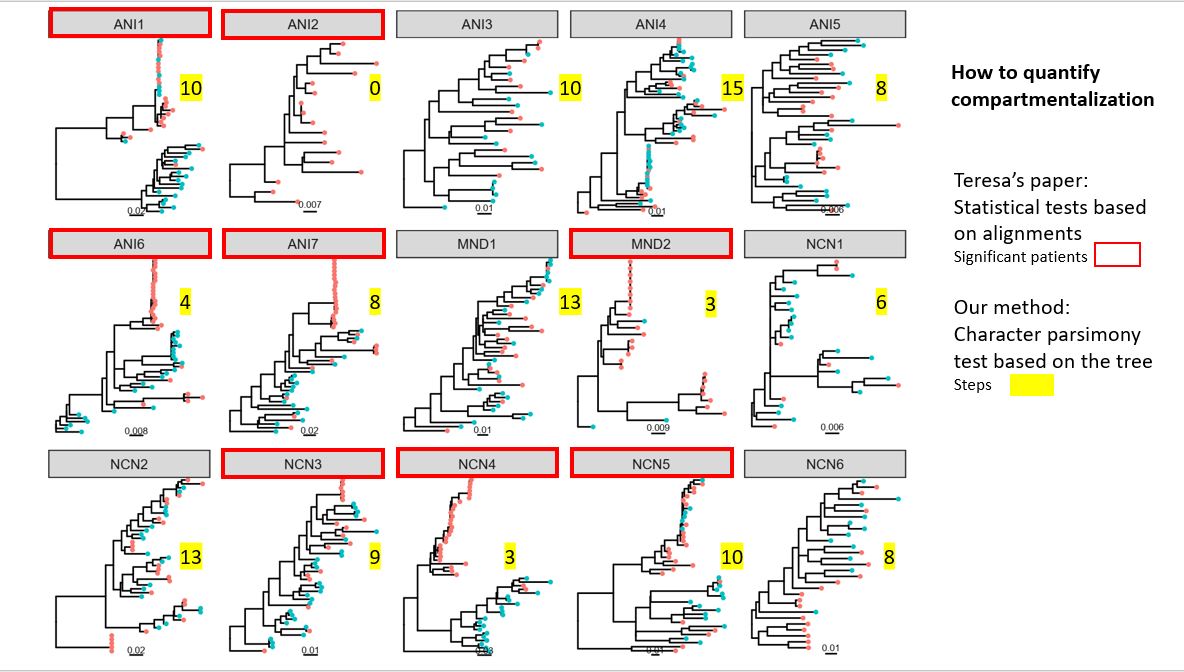
[[File:HIV-compartmentalization.jpg|thumbnail|by Lily]] | |||
* Participants: Lily | * Participants: Lily | ||
* Questions and goals | * Questions and goals | ||
| Line 44: | Line 96: | ||
* Approach | * Approach | ||
** Evolutionary mechanisms: mutation, recombination, and adaptive selection | ** Evolutionary mechanisms: mutation, recombination, and adaptive selection | ||
** Homoplasy index as a measure of compartmentalization? | ** Homoplasy index as a measure of compartmentalization? Randomization to obtain p-values of HI. | ||
** Evolutionary rates & signature (BEAST) | ** Evolutionary rates & signature (BEAST) | ||
** Tests of natural selection (PAML, MK analysis) | ** Tests of natural selection (PAML site models, branch-site models & MK analysis) | ||
** Phylogenetic analysis: tree per individual; supertree; haplotype networks (per individual) | |||
** Simulated compartmentalization | |||
==Project 4. Natural Language models of proteins== | ==Project 4. Natural Language models of proteins== | ||
| Line 54: | Line 108: | ||
# Fine-tuning for OspC, to be integrated with the centroid algorithm | # Fine-tuning for OspC, to be integrated with the centroid algorithm | ||
# 2nd-generation centroid design: k-means algorithm (with applications to vls, Dengue, flu B) | # 2nd-generation centroid design: k-means algorithm (with applications to vls, Dengue, flu B) | ||
* Reading list | * Tools & Reading list | ||
** [https://github.com/facebookresearch/esm Facebook ESM: pre-trained language models (for feature extraction)] | |||
*** Step 1. implementation with colab | |||
*** Step 2. Fine-tuning with OspC seqs; extract embedding | |||
*** Step 3. Applications: classify (OspC vs VlsE), contact map (native vs synthetics), solubility | |||
** Strodthoff et al (2020). Bioinformatics. [https://academic.oup.com/bioinformatics/article/36/8/2401/5698270 UDSMProt: universal deep sequence models for protein classification]. [https://github.com/nstrodt/UDSMProt Source code on Github] | ** Strodthoff et al (2020). Bioinformatics. [https://academic.oup.com/bioinformatics/article/36/8/2401/5698270 UDSMProt: universal deep sequence models for protein classification]. [https://github.com/nstrodt/UDSMProt Source code on Github] | ||
** [https://www.pnas.org/content/118/15/e2016239118 Rives et al (2021). PNAS. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences.] [https://github.com/facebookresearch/esm Github repository] | ** [https://www.pnas.org/content/118/15/e2016239118 Rives et al (2021). PNAS. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences.] [https://github.com/facebookresearch/esm Github repository] | ||
** Transformer: [https://arxiv.org/pdf/1706.03762.pdf Vaswani et al (2017). Attention is All You Need] [https://github.com/huggingface/transformers Github repository for Huggingface/Transformer] | |||
Latest revision as of 13:49, 24 August 2021
Group meeting/Field trip schedule
August 2021
- Wrapping up: Bb transcriptomes, make interactive plots using ggplotly. Interactive volcano plots:
- Starvation response: differentially expressed genes. Work credit (data collection, R analysis, and web-interactive visualization): Hannah Ford. Data source: Drecktah et al (2015). PLoS Pathogen
- Borrelia starvation response: differentially expressed sRNA. Work credit: Jackie Yee. Data source: Drecktrah et al (2018).
- Response to Doxycycline treatment. Work credit: Jacki Yee. Data source: Caskey et al (2019)
- Monte Carlo Club Season IV. Machine learning with Scikit-Learn
- First publication on computational vaccine design (with a blog post): Jenner's Dilemma.
July 2021
- Belfer housekeeping duties: During the month of July, members of Qiu lab (including the PI) will be responsible for the following:
- Performing a careful sweep of the common seating, eating, and food preparation areas each Monday, Wednesday, and Friday. The purpose of this sweep is to do some light organization/cleaning and to identify persistent cleanliness problems that should then be reported (via the lab’s PI) to myself and Elizabeth Cohn (ec2692@hunter.cuny.edu), and the collective PIs of the floor.
- Throwing away any personal food (and personal food containers) left in the refrigerators beyond 5 pm on each Friday.
- Keeping general tabs on the bathrooms of the floor to ensure cleanliness. Persistent cleanliness problems that are unreasonable for the WCMC custodial staff should be addressed by the members of the floor.
- July 2, Friday. Field trip
- location: Rockefeller State Park
- Participants: Saymon, Lia, Joy, Weigang
- Lab meeting: Weekly on Tuesday 11-1
June 2021
- June 3, 2021 (Thursday). Summer research kickoff
- June 8, 2021 (Tuesday). 11-2
- Algorithm development (Brian)
- NLP models of protein structure (Eamen, Roman, Edgar)
- Bb transcriptomics (Niemah & Jackie)
- HIV compartmentalization (Lily)
- June 10, 2021 (Thursday). No meeting. Field day
- Location: Hackscher State Park, Long Island
- Participants: Desiree, Lily, Lia, John, Weigang
- Outcome: ~60 nymph ticks
- June 15, 2021 (Tuesday). 11-2. Lab meeting
- June 17, 2021 (Thursday). No meeting. Field day
- Location: Quabbin Reservoir, MA
- Participant: Saymon, Brian, Lia, Weigang
- Outcome: ~130 nymph ticks
- June 22, 2021 (Tuesday). 11-1
- Niemah & Jackie will work on collecting transcriptome data into Excel sheets
- Lily presented latest trees on HIV compartmentalization
- Roman will work on env gene embedding using Facebook pretrained ESM model
- June 24, 2021 (Thursday). Lab meeting 11-1
- June 30, 2021 (Wed). Lab meeting 11-1
Project 1. Borrelia genomics
- Participants: Niemah, Jackie
- Questions & Goals:
- Upgrade database, genome pipeline, and website (Lia)
- Phylogeography & evolutionary maintenance of divided genome (Saymon)
- vls evolution (with simulation) & development of immunoflorescence microsopy methods(Lily). Live imaging.
- Reading list
- Latest review book Lyme Disease and Relapsing Fever Spirochetes: Genomics, Molecular Biology, Host Interactions and Disease Pathogenesis. The chapter on gene regulation and transcriptomics (notice Fig 1, Fig 2, and Table 1)
- Schward et al (2021). Multipartite Genome of Lyme Disease Borrelia: Structure, Variation and Prophages
- Stevenson & Seshu (2018). Regulation of Gene and Protein Expression in the Lyme Disease Spirochete
- The bowtie model of gene regulation: Yan et al (2017). PLoS Comp Biol.
Project 2. Design algorithms for vaccines
- Participants: Dr Saad Mneimneih (CS Department), Brian
- Questions & Goals:
- Generalized algorithms for antigen with arbitrary tree shape
- Scikit-learn K-means
- Data set 1. Neutral evolution (with exponentially distributed branch lengths). Binary strings (L=100 bits) evolved from a coalescent tree of 20 leaves. Simulated with
rcoal(20); rTraitDisc; simSeq(). code from previous work - Data set 2. Two major clades. HA sequences from fluB
- Data set 3. Four major clades. Dengue
- Data set 4. Star-shaped tree, driven by recombination. OspC
- Data set 5. Multiple major clades. vls cassette in Lyme species
- Combination algorithms
- Naive Bayes models to integrate immunogenicity data
- Natural language models to improve structural stability (see Project 4 below)
- Improve protein solubility with Solubis
- Estimating Ag-Ab binding parameters with hierarchical Bayesian modeling:
- Generalized algorithms for antigen with arbitrary tree shape
- Reading list
- Greet De Baets, Joost Van Durme, Rob van der Kant, Joost Schymkowitz, Frederic Rousseau, Solubis: optimize your protein, Bioinformatics, Volume 31, Issue 15, 1 August 2015, Pages 2580–2582, https://doi.org/10.1093/bioinformatics/btv162
- Degoot et al (2019). Predicting Antigenicity of Influenza A Viruses Using biophysical ideas
- Di et al (2021). Maximum antigen divergence in Lyme bacterial population
- Golovanov et al (2004). A Simple Method for Improving Protein Solubility and Long-Term Stability. J. Am. Che. Soc.
Project 3. HIV compartmentalized evolution
- Participants: Lily
- Questions and goals
- Do HIV evolve cell type tropisms within the host? Specifically, the Neural(N)-tropism vs T-cell(T)-tropism?
- Build a classifier of N-tropism HIV subtypes
- A presentation for an HIV conference in October
- Reading list
- HIV compartmentalized evolution: Evering et al (2014)
- Data sets
- ~500 sequences of env genes from 15 patients
- 2nd time point single-cell genome sequences for some of the patients
- Experimentally verified N-tropism subtypes
- Approach
- Evolutionary mechanisms: mutation, recombination, and adaptive selection
- Homoplasy index as a measure of compartmentalization? Randomization to obtain p-values of HI.
- Evolutionary rates & signature (BEAST)
- Tests of natural selection (PAML site models, branch-site models & MK analysis)
- Phylogenetic analysis: tree per individual; supertree; haplotype networks (per individual)
- Simulated compartmentalization
Project 4. Natural Language models of proteins
- Participants: Eamen, Roman, and Edgar
- Questions & Goals
- Learn, implement, and compare the existing tools
- Fine-tuning for OspC, to be integrated with the centroid algorithm
- 2nd-generation centroid design: k-means algorithm (with applications to vls, Dengue, flu B)
- Tools & Reading list
- Facebook ESM: pre-trained language models (for feature extraction)
- Step 1. implementation with colab
- Step 2. Fine-tuning with OspC seqs; extract embedding
- Step 3. Applications: classify (OspC vs VlsE), contact map (native vs synthetics), solubility
- Strodthoff et al (2020). Bioinformatics. UDSMProt: universal deep sequence models for protein classification. Source code on Github
- Rives et al (2021). PNAS. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Github repository
- Transformer: Vaswani et al (2017). Attention is All You Need Github repository for Huggingface/Transformer
- Facebook ESM: pre-trained language models (for feature extraction)