NYRaMP-Informatics-2024: Difference between revisions
(Created page with "<center>'''BioMedical Genomics'''</center> <center>August 2024, Tuesdays 10-12 noon, DNA Learning Center</center> <center>'''Instructor:''' Weigang Qiu, Ph.D., Professor, Department of Biological Sciences, Hunter College, CUNY; '''Email:''' wqiu@.hunter.cuny.edu</center> <center>'''T.A.:''' Brandon Ely, CUNY Graduate Center; '''Email:bely@gradcenter.cuny.edu'''</center> <center> {| class="wikitable" |- ! MA plot !! Volcano plot !! Heat map |- | File:GeneExp1.jpeg|300px...") |
|||
| (53 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<center>''' | <center>'''NYRaMP Informatics Workshop'''</center> | ||
<center>August 2024, Tuesdays 10-12 noon, DNA Learning Center</center> | <center>August 2024, Tuesdays 10-12 noon, DNA Learning Center</center> | ||
<center>''' | <center>'''Instructors:''' Weigang Qiu (Professor, Hunter College, wqiu@.hunter.cuny.edu) & Brandon Ely (CUNY Graduate Center, bely@gradcenter.cuny.edu)</center> | ||
<center> | <center> | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 14: | Line 13: | ||
|} | |} | ||
</center> | </center> | ||
== | ==Overview== | ||
A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and rapid DNA and RNA-sequencing technologies, biomedical sciences are undergoing a rapid & irreversible transformation into a highly data-intensive field, that requires familiarity with concepts in both biological, computational, and statistical sciences. | |||
Genome information is revolutionizing virtually all aspects of life sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as statistics. | Genome information is revolutionizing virtually all aspects of life sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as statistics. | ||
This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises | This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises. | ||
==Learning goals== | ==Learning goals== | ||
By the end of this | By the end of this workshop students will be able to: | ||
* | * Visualize genomics data using R & RStudio | ||
* | * Perform t-tests, regression analysis, and association tests with R & Rstudio | ||
* Interpret results of data visualization & statistical analysis | |||
* | |||
==Web Links== | ==Web Links== | ||
* | * Cloud R account (free): https://posit.cloud/; Join the shared work space "NYRaMP-Informatics" | ||
* | * For your own computer, download the desktop version: https://posit.co/download/rstudio-desktop/ | ||
* Textbook: [http://r4all.org/#about Introduction to R for Biologists] | * Textbook: [http://r4all.org/#about Introduction to R for Biologists] | ||
* Download: [http://www.r4all.org/books/datasets R datasets] | * Download: [http://www.r4all.org/books/datasets R datasets] | ||
* A reference book: [https://r4ds. | * A reference book: [https://r4ds.hadley.nz/ R for Data Science (Wickharm & Grolemund)] | ||
==Week 1. Aug 6== | |||
* Pre-test: visualization, interpretation, and stats. Download file: [[File:Pre-test.pdf|thumb]] | |||
* Computer/Cloud setup & software download/installation | |||
* R Tutorial 1. Getting started: Basics: interface, packages, variables, objects, functions. Download slides: [[File:R-tutorials-Part-1.pdf|thumb]] | |||
* Practice #1 | |||
==Week 2. Aug 13== | |||
* R Tutorial 2. Data manipulation. Download slides: [[File:R-tutorials-Part-2.pdf|thumb]] | |||
* Practice #2 | |||
==Week 3. Aug 20== | |||
* R Tutorial 3. Data visualization. Lecture slides: [[File:R-tutorials-Part-3.pdf|thumb]] | |||
* Practice #3 | |||
==Week 4. Aug 27== | |||
* Tutorial #4. t-test and regression analysis. Slides: [[File:R-tutorials-Part-4.pdf|thumb]] | |||
* Practice #4 | |||
Advanced topics (for Self Study) | |||
* R Demo I: [https://borreliabase.org/~wgqiu/tutorial-markdown.html#part-1.-t-test-for-two-groups Documentation & presentation with R Markdown (Part I)] | |||
* R Demo II: [https://borreliabase.org/~wgqiu/r-demo-2024.html#part-4.-advanced-topic-cluster-analysis-of-multi-dimensional-data Cluster analysis (Part 4)] | |||
* R Demo III: [https://borreliabase.org/~wgqiu/r-demo-2024.html#part-5.-case-study-differential-gene-expressions-in-lyme-diease-bacteria Differential gene expression analysis (Part 5)] | |||
Latest revision as of 01:44, 27 August 2024
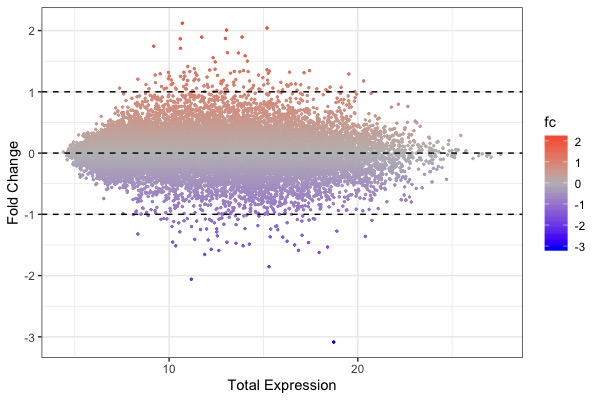
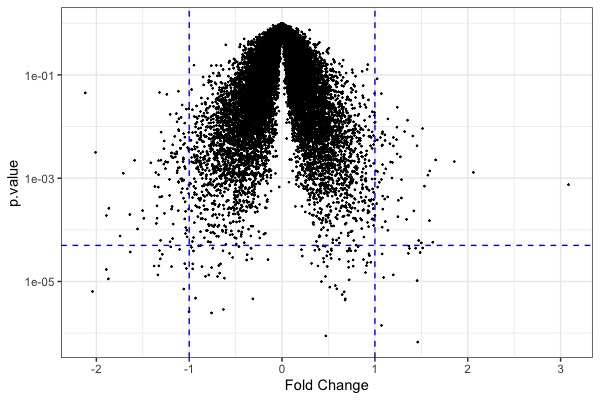
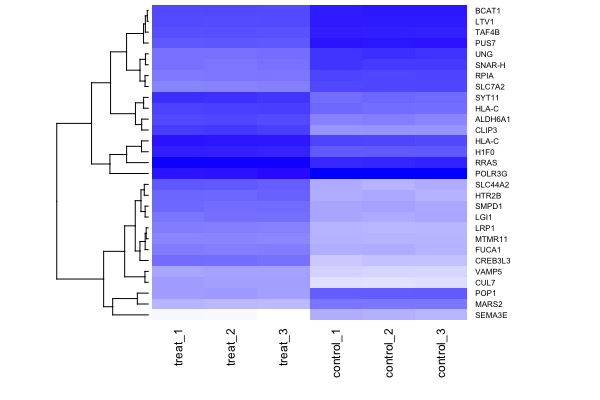
| MA plot | Volcano plot | Heat map |
|---|---|---|
Overview
A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and rapid DNA and RNA-sequencing technologies, biomedical sciences are undergoing a rapid & irreversible transformation into a highly data-intensive field, that requires familiarity with concepts in both biological, computational, and statistical sciences.
Genome information is revolutionizing virtually all aspects of life sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as statistics.
This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises.
Learning goals
By the end of this workshop students will be able to:
- Visualize genomics data using R & RStudio
- Perform t-tests, regression analysis, and association tests with R & Rstudio
- Interpret results of data visualization & statistical analysis
Web Links
- Cloud R account (free): https://posit.cloud/; Join the shared work space "NYRaMP-Informatics"
- For your own computer, download the desktop version: https://posit.co/download/rstudio-desktop/
- Textbook: Introduction to R for Biologists
- Download: R datasets
- A reference book: R for Data Science (Wickharm & Grolemund)
Week 1. Aug 6
- Pre-test: visualization, interpretation, and stats. Download file: File:Pre-test.pdf
- Computer/Cloud setup & software download/installation
- R Tutorial 1. Getting started: Basics: interface, packages, variables, objects, functions. Download slides: File:R-tutorials-Part-1.pdf
- Practice #1
Week 2. Aug 13
- R Tutorial 2. Data manipulation. Download slides: File:R-tutorials-Part-2.pdf
- Practice #2
Week 3. Aug 20
- R Tutorial 3. Data visualization. Lecture slides: File:R-tutorials-Part-3.pdf
- Practice #3
Week 4. Aug 27
- Tutorial #4. t-test and regression analysis. Slides: File:R-tutorials-Part-4.pdf
- Practice #4
Advanced topics (for Self Study)
- R Demo I: Documentation & presentation with R Markdown (Part I)
- R Demo II: Cluster analysis (Part 4)
- R Demo III: Differential gene expression analysis (Part 5)