Summer 2018: Difference between revisions
imported>Weigang m (→Journal Club) |
imported>Lia |
||
| (78 intermediate revisions by 3 users not shown) | |||
| Line 13: | Line 13: | ||
# Dr Lia Di, Senior Scientist | # Dr Lia Di, Senior Scientist | ||
# Dr Weigang Qiu, Principal Investigator | # Dr Weigang Qiu, Principal Investigator | ||
# Summer Interns | # Summer Interns: Muhammad, Pavan, Roman, Benjamen, Andrew, Michelle, Hannah | ||
=Journal Club= | =Journal Club= | ||
# [http://diverge.hunter.cuny.edu/labwiki/First_Time_Guide#A_UNIX_.26_Perl_Primer a Unix & Perl tutorial] | # [http://diverge.hunter.cuny.edu/labwiki/First_Time_Guide#A_UNIX_.26_Perl_Primer a Unix & Perl tutorial] | ||
# A short introduction to molecular phylogenetics: http://www.ncbi.nlm.nih.gov/pubmed/12801728 | # A short introduction to molecular phylogenetics: http://www.ncbi.nlm.nih.gov/pubmed/12801728 | ||
# A review on Borrelia genomics: https://www.ncbi.nlm.nih.gov/pubmed/24704760 | # A review on Borrelia genomics: https://www.ncbi.nlm.nih.gov/pubmed/24704760 | ||
# ospC epitope mapping: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0067445 | |||
# Codon usage changes fitness in E.coli: [http://www.pnas.org/content/115/21/E4940.long Frumkin et al (2018) "Codon usage of highly expressed genes affects proteome-wide translation efficiency". PNAS] | |||
=Projects= | =Projects= | ||
==Borrelia genome evolution (Led by Saymon)== | ==Borrelia genome evolution (Led by Saymon)== | ||
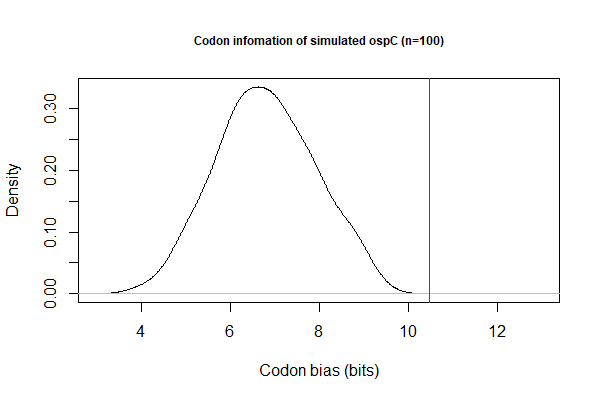
[[File:Codon-info1.png|thumbnail|Codon biase measured by Shanon information (bits)]] | |||
# Goal 1. Estimate time of cross-Atlantic dispersal using core-genome sequences | # Goal 1. Estimate time of cross-Atlantic dispersal using core-genome sequences | ||
# Goal 2. Investigate codon biases with respect to levels of gene expression | # Goal 2. Investigate codon biases with respect to levels of gene expression. Data file: [[File:B31-cp26.txt|thumbnail]] | ||
<b>* Andrew's BioPython code to calculate CAI</b> | |||
<syntaxhighlight lang="python"> | |||
#Opens the fasta file and reads contents into a string (myStr). | |||
myFile = open("B31-cp26.txt","r") | |||
myStr = myFile.read() | |||
myFile.close() | |||
#Imports codon usage module. | |||
from Bio.SeqUtils import CodonUsage as cu | |||
#Takes myStr and processes it into a list of sequences (FastaList). | |||
FastaList = myStr.split(">") | |||
FastaList = FastaList[1:] | |||
IDList = [] | |||
EnterList = [] | |||
##Separates FastaList into a list of sequence IDs (IDList) and a list of sequences (EnterList). | |||
for seq in FastaList: | |||
IDList += [seq[:6]] | |||
EnterList += [seq[6:]] | |||
##Removes enter characters from each sequence in EnterList. | |||
SeqList = [] | |||
for seq in EnterList: | |||
SeqStr = seq.replace("\n", "") | |||
SeqList += [SeqStr] | |||
#Calculates and presents the CAI value for each sequence using functions from the module. | |||
myObject = cu.CodonAdaptationIndex() | |||
myObject.generate_index("B31-cp26.txt") | |||
for SeqIndex in range(len(SeqList)): | |||
print (IDList[SeqIndex], ' CAI =', myObject.cai_for_gene(SeqList[SeqIndex])) | |||
</syntaxhighlight> | |||
Output for cp26:<br> | |||
BB_B01 CAI = 0.7190039074113422<br> | |||
BB_B02 CAI = 0.678404951527374<br> | |||
BB_B03 CAI = 0.6893076488255271<br> | |||
BB_B04 CAI = 0.7250154635421513<br> | |||
BB_B05 CAI = 0.6971190458423587<br> | |||
BB_B06 CAI = 0.67042305582205<br> | |||
BB_B07 CAI = 0.6971020959083346<br> | |||
BB_B09 CAI = 0.6786931743972611<br> | |||
BB_B10 CAI = 0.7224886929887183<br> | |||
BB_B12 CAI = 0.6997502136447451<br> | |||
BB_B13 CAI = 0.7592966148479222<br> | |||
BB_B14 CAI = 0.6959525612884284<br> | |||
BB_B16 CAI = 0.6835709626613392<br> | |||
BB_B17 CAI = 0.6974779110749645<br> | |||
BB_B18 CAI = 0.7052250722958308<br> | |||
BB_B19 CAI = 0.7049049245887261<br> | |||
BB_B22 CAI = 0.6860641572293008<br> | |||
BB_B23 CAI = 0.6915165725213809<br> | |||
BB_B24 CAI = 0.7025276490965267<br> | |||
BB_B25 CAI = 0.7439914547011712<br> | |||
BB_B26 CAI = 0.7255623088410704<br> | |||
BB_B27 CAI = 0.7161378416520467<br> | |||
BB_B28 CAI = 0.7316661839512337<br> | |||
BB_B29 CAI = 0.6919705705489939<br> | |||
# Codon bias by Shannon information: BASH pipeline | |||
## Simulate 100 sequences for each CDS: <code>cat filename.txt | while read line; do ./codon-info-sim.pl -n 100 BbB31.cutg.GCG "$line".fas > "$line"-sim.fas; done; </code> | |||
## Calculate Shannon index for each CDS (simulated & actual)<code>./codon-info.pl BbB31.cutg.GCG foo.fas</code> | |||
==Identification of host species from ticks (Led by Lily [after first-level])== | ==Identification of host species from ticks (Led by Lily [after first-level])== | ||
# Goal 1. Protocol optimization for PCR amplification of host DNA from ticks | # Goal 1. Protocol optimization for PCR amplification of host DNA from ticks | ||
| Line 30: | Line 94: | ||
# Goal 1. Association of genes/SNPs with biofilm formation and c-di-GMP levels: Manuscript preparation | # Goal 1. Association of genes/SNPs with biofilm formation and c-di-GMP levels: Manuscript preparation | ||
# Goal 2. Association of genome diversity with metabolic diversity | # Goal 2. Association of genome diversity with metabolic diversity | ||
* (Christopher) This script parses excel peak-area file into database & R inputs | |||
<syntaxhighlight lang="perl"> | |||
#!/usr/bin/perl -w | |||
use strict; | |||
use Data::Dumper; | |||
use Getopt::Std; | |||
my %opts; | |||
my $line_ct = 0; | |||
my (@colnames, @areas, %seen_cmps, %seen_gids); | |||
getopts('dr', \%opts); | |||
while(<>) { | |||
chomp; | |||
$line_ct++; | |||
next unless $line_ct >=4; | |||
if ($line_ct == 4) { | |||
@colnames = split "\t", $_; | |||
for (my $i=5; $i<=$#colnames; $i++) { $seen_gids{$colnames[$i]}++ } # get uniq gids | |||
next; | |||
} | |||
my @data = split "\t", $_; | |||
$seen_cmps{$data[1]}++; # get unique compound formula | |||
for (my $i=5; $i<=$#data; $i++) { | |||
my $area = { 'compound' => $data[1], 'gid' =>$colnames[$i], 'peak_area' => $data[$i]}; | |||
push @areas, $area; | |||
} | |||
} | |||
if ($opts{d}) { # for database output | |||
foreach my $cmp (sort keys %seen_cmps) { | |||
foreach my $gid (sort keys %seen_gids) { | |||
my @peaks = grep { $_->{compound} eq $cmp && $_->{gid} == $gid } @areas; | |||
my $peak_str = join ",", map {$_->{peak_area} || "NULL"} @peaks; | |||
print join "\t", ($gid, $cmp, "{" . $peak_str . "}"); | |||
print "\n"; | |||
} | |||
} | |||
} | |||
if ($opts{r}) { # for R output | |||
foreach my $cmp (sort keys %seen_cmps) { | |||
foreach my $gid (sort keys %seen_gids) { | |||
my @peaks = grep { $_->{compound} eq $cmp && $_->{gid} == $gid } @areas; | |||
foreach my $peak (@peaks) { | |||
next unless $peak->{peak_area}; | |||
print join "\t", ($peak->{peak_area}, $gid, $cmp); | |||
print "\n"; | |||
} | |||
} | |||
} | |||
} | |||
exit; | |||
</syntaxhighlight> | |||
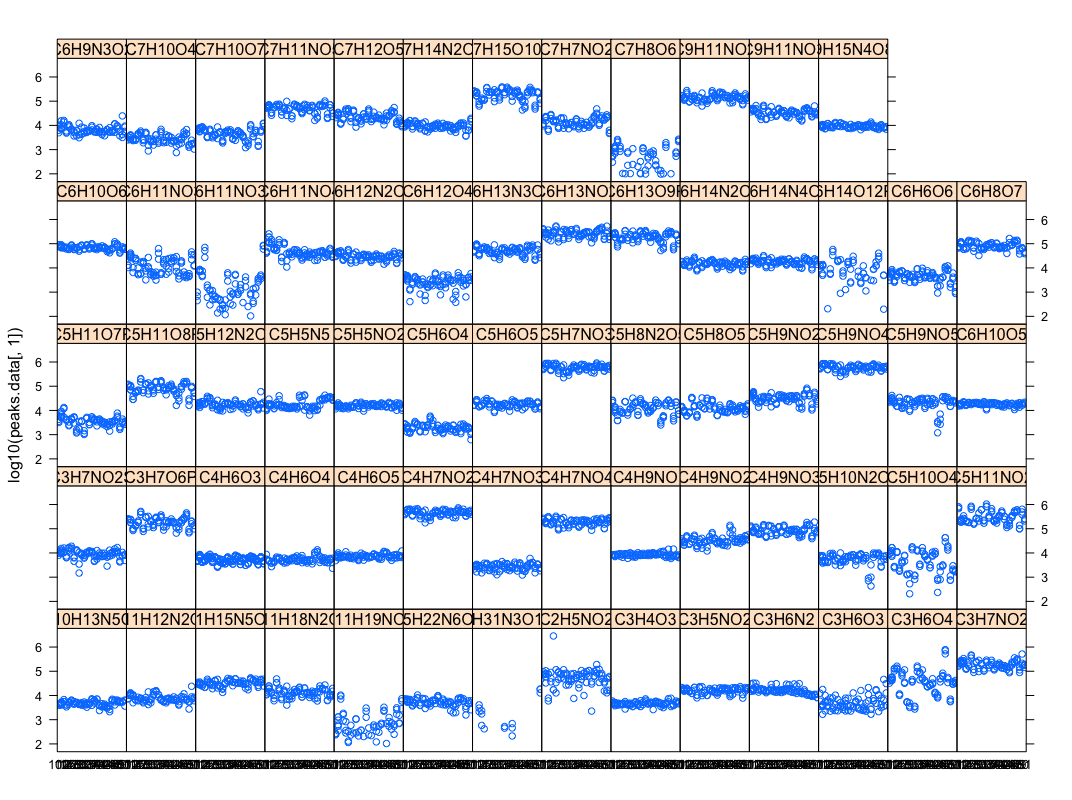
[[File:Stripplot-peaksdatabycompound.png|thumbnail|Compound amount in each genome]] | |||
*(Benjamen) Creates a sample simple force directed network in R using networkD3. Install packages "igraph" and "networkD3". | |||
[https://benjilev.shinyapps.io/simplesample/ Simple Sample] | |||
<syntaxhighlight lang="Bash"> | |||
library(igraph) | |||
library(networkD3) | |||
nodes <- c(rep('x1', 10), rep('x2',10), rep('x3',10), rep('x4',10)) | |||
ids <- c(1,1,1,-1,0,-1,0,0,-1,0,0,0,0,0,0,0,0,0,1,-1,0,0,0,0,0,1,-1,-1,0,0,0,0,0,1,-1,0,0,0,0,0) | |||
df <- data.frame(nodes, ids) | |||
df$ids | |||
matrix(df$ids, nrow = 4, ncol = 10, byrow = T) | |||
testmat<-matrix(df$ids, nrow = 4, ncol = 10, byrow = T) | |||
rownames(testmat) <- c("x1", "x2", "x3", "x4") | |||
colnames(testmat) <- c("v1", "v2", "v3", "v4", "v5", "v6", "v7", "v8", "v9", "v10") | |||
testmat.inc <- graph_from_incidence_matrix(testmat) | |||
testmat.edge <- as_edgelist(testmat.inc) | |||
colnames(testmat.edge) <- c("From", "To") | |||
testmat.df <- as.data.frame(testmat.edge) | |||
simpleNetwork(testmat.df, opacity = .8, fontSize = 16) | |||
</syntaxhighlight> | |||
*(Benjamen) Force-Directed Network Graph Visualization of the iJN746 S Matrix (Only Metabolites in Our Database) | |||
[http://rpubs.com/benjilev/NetworkModel/ Force-Directed Network Graph of iJN746 (Only Metabolites in Our Database)] | |||
<syntaxhighlight lang="Bash"> | |||
library(igraph) | |||
library(networkD3) | |||
#create a preliminary edgelist from the original matrix. If using the simpleNetwork() function, this | |||
#is the only required step. | |||
stwo <- read.table("Stwo.tsv", header=T) | |||
stwo.inc <- graph_from_incidence_matrix(stwo) | |||
stwo.edge <- as_edgelist(stwo.inc) | |||
colnames(stwo.edge) <- c("from", "to") | |||
stwo.df <- as.data.frame(stwo.edge) | |||
#both the "from" and "to" columns must be given a unique numerical value, as this is the only way | |||
#forceNetwork() will run. To assign numerical values, we index each unique instance of a metabolite | |||
#or reaction in order. | |||
#index "from" column | |||
unique(stwo.edge[,1]) | |||
order(unique(stwo.edge[,1])) | |||
from.idx <- data.frame(row.names=unique(stwo.edge[,1]), row.idx=1:82) | |||
stwo.df$from | |||
from.idx[as.character(stwo.df$from),1] | |||
stwo.df$from.idx <- from.idx[as.character(stwo.df$from),1] | |||
#index "to" column | |||
unique(stwo.edge[,2]) | |||
order(unique(stwo.edge[,2])) | |||
to.idx <- data.frame(row.names=unique(stwo.edge[,2]), row.idx=1:188) | |||
stwo.df$to | |||
to.idx[as.character(stwo.df$to),1] | |||
stwo.df$to.idx <- to.idx[as.character(stwo.df$to),1] | |||
#add 81 to every value in "to" column and subtract 1 from every value in "from" column. This is so | |||
#the indexing starts from 0, and every indexed variable on the "to" side continues from where "from" | |||
#left off, which, in this case, is variable #82. | |||
stwo.df$to.idx <- stwo.df[,4]+81 | |||
stwo.df$from.idx <- stwo.df[,3]-1 | |||
#create new data frame with just the "from" and "to" columns. | |||
stwo.new <- data.frame(stwo.df$from.idx, stwo.df$to.idx) | |||
colnames(stwo.new) <- c("from", "to") | |||
#order the edgelist by the to column. The ordered "to" column is then appended to the "from" column, | |||
#and the now disordered "from" column is appended to the "to" column, effectivley mirroring the original | |||
#edgelist. This is done because every variable in the network must show up in the "from" column for | |||
#the forceNetwork() function to work. | |||
stwo.new2 <- stwo.new[order(stwo.new$to),] | |||
#zoom in and out by rolling mouse wheel or double clicking. Click and drag to pan the model. | |||
forceNetwork(Links = links, Nodes = nodes, Source="from", Target = "to", NodeID = "idn", Group = "type.label", | |||
linkWidth = 1, linkColour = "green", fontSize = 20, zoom = T, legend = T, Nodesize = 6, opacity = 1, | |||
charge = -50, width = 1920, height = 1080) | |||
</syntaxhighlight> | |||
==Machine learning approaches to evolution (Led by Oliver & Brian)== | ==Machine learning approaches to evolution (Led by Oliver & Brian)== | ||
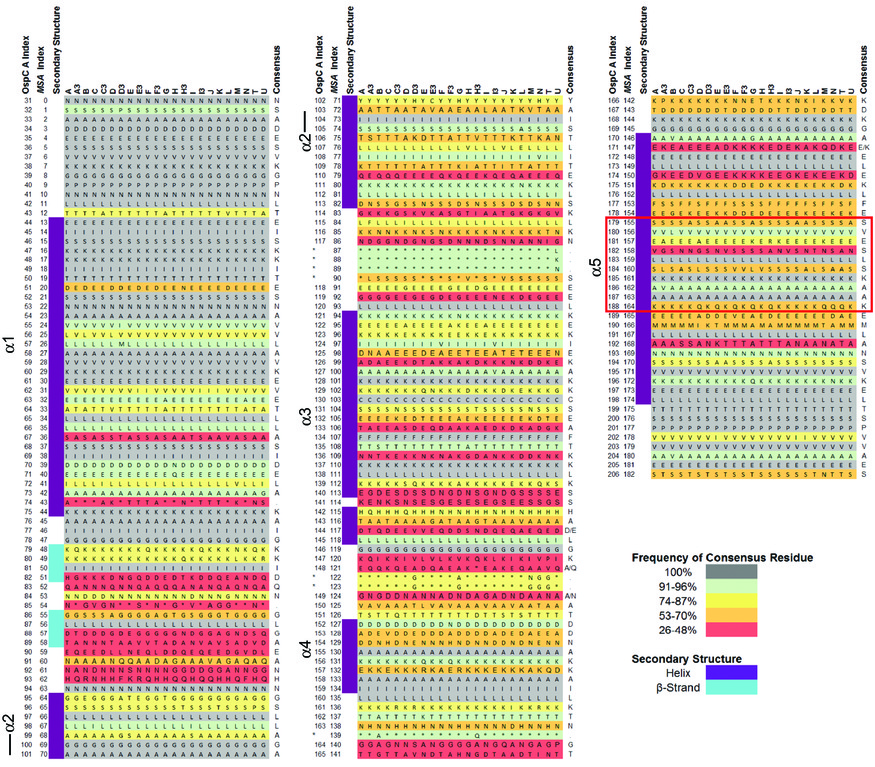
[[File:S2-lo-res0.jpg|thumbnail|OspC structural alignment, converted from S2 from Baum et al (2013)]] | |||
# Goal 1. Implement Hopfield network for optimization of protein structure | # Goal 1. Implement Hopfield network for optimization of protein structure | ||
# Goal 2. Neural-net models of OspC | # Goal 2. Neural-net models of OspC. Structural alignment (S2 from Baum et al 2013): | ||
# Goal 3. K-mer-based pipeline for genome classification | # Goal 3. K-mer-based pipeline for genome classification | ||
=Schedule= | =Weekly Schedule= | ||
* Summer kickoff (June 1, 2018, Friday): Introduction & schedules | |||
* June | * Week 1 (June 4-8): | ||
* June | ** Monday: [http://diverge.hunter.cuny.edu/labwiki/First_Time_Guide#A_UNIX_.26_Perl_Primer the Unix & Perl Tutorial], Part 1 | ||
** Tuesday: Unix Part2. Explore the "iris" data set using R, by following the [[Monte_Carlo_Club#Season_II._Summer_2017_.28Theme:_Machine_Learning.29|the Monte Carlo Club Week 1 (1 & 2) and Week 2]]. Read McKay (2003), Chapters 38 & 39 | |||
* | ** Thursday: 1st field day (Caumsett State Park); Participants: John, Muhammad, Pavan, Andrew, Dr Sun, Weigang, with 3 members of Moses team from Suffolk County Vector Control. Got ~110 deer tick nymphs | ||
* June | ** Friday: meeting with MSKCC group at 11am; BBQ afterwards | ||
* | * Week 2 (June 11-15): | ||
* | ** Monday: Lab meeting, projects assigned | ||
* | ** Tuesday: neural net tutorial (by Brian) | ||
* July | ** Thursday: 2nd field day (Fire Island National Seashore). Participants: John, Brian, Mei, Muhammad, Pavan, Benjamin, and Weigang. Got ~100 lone-star ticks and 4 deer tick nymphs | ||
* July 12 ( | * Week 3 (June 18-22): | ||
* | ** Monday: Lab meeting, 1st project reports | ||
* | *** Codon Bias: Theory, Coding, and Data (Andrew, Pavan, Saymon) | ||
*** OspC epitope identification: Serum correlation, sequence correlation, immunity-sequence correction (Muhammad, Roman, Brian) | |||
*** Pseudomonas metabolomics: parsing intensity file; theory & parsing SMBL file (Chris & Benjamin) | |||
** Tuesday: working groups | |||
** Wed: working groups | |||
** Thursday: Big Data Workshop | |||
** Friday: working groups | |||
* Week 4 (June 25-29): | |||
** Monday: Lab meeting | |||
** Tu-Th: work sessions | |||
** Friday: Joint lab meeting with MSKCC/Lab visit | |||
* Week 5 (July 2-6): | |||
** Monday: Lab meeting | |||
** Tuesday/Wed: 4th of July Break | |||
** Thursday & Friday: work sessions | |||
* Week 6 (July 9-13) | |||
** Monday (July 9): E-reports & Wiki posts | |||
* Week 7 (July 16-20) | |||
** Monday (July 16): E-reports * Wiki posts | |||
* Week 8 (July 23-27) | |||
** Monday: lab meeting resumes | |||
** Monte Carlo Club, Season 3 (Evolutionary Computing) starts | |||
* Week 9 (July 30-Aug 3) | |||
* Week 10 (Aug 6-Aug 10) | |||
** Final report | |||
=Lab notes for Summer HS Interns= | |||
* NCI Cloud: [http://www.cancergenomicscloud.org/ Seven Bridges Cloud Platform]. Create an user account | |||
* Read documentation & tutorials: [https://docs.cancergenomicscloud.org/docs Documentation] | |||
=Notes & Scripts= | |||
* (Weigang) Re-analysis of ospC cross-reactivity using normalization: | |||
** Fold change vs alleles for mouse + humans: [http://diverge.hunter.cuny.edu/~weigang/heatmap-total.html interactive heatmap by heatmaply] | |||
** Correlation matrix based on N=23 mouse samples (and 7 controls): [http://diverge.hunter.cuny.edu/~weigang/cor-test-mouse.html interactive heatmap by heatmaply] | |||
** Correlation matrix based on N=55 human samples (and 25 controls): [http://diverge.hunter.cuny.edu/~weigang/cor-test-human.html interactive heatmap by heatmaply] | |||
** Correlation matrix based on both mouse & human samples: [http://diverge.hunter.cuny.edu/~weigang/cor-test-total.html interactive heatmap by heatmaply]] | |||
* (Weigang) A sample R script to parse [https://doi.org/10.1371/journal.pone.0067445.s006 Table S2 from Baum et al 2013], sera-antigen reactivity measurements | |||
<syntaxhighlight lang="bash"> | |||
# preliminaries: save as TSV; substitute "\r" if necessary; | |||
# substitute "N/A" to "NA"; remove extra columns | |||
setwd("Downloads/") | |||
x <- read.table("table-s2.txt4", sep="\t", header=T) | |||
View(x) | |||
colnames(x) | |||
which(x[,8]=="A") | |||
x[which(x[,8]=="A"),12] | |||
x[which(x[,8]=="A3"),12] | |||
cor.test(x[which(x[,8]=="A3"),12], x[which(x[,8]=="A"),12], method = "pearson") | |||
x.cor$estimate | |||
levels(x[,8]) # obtain ospC allele types; to be looped through in pairwise | |||
for (i in 1:?) { for (j in ?:?) {cor.test(....)}} | |||
</syntaxhighlight> | |||
* (Muhammad) Output generates data frame of correlation/p values for 23 different Osp-C allele types in pairwise | |||
<syntaxhighlight lang="bash"> | |||
setwd("C:/R_OspC") | |||
x <- read.table("Table-S2.txt", sep="\t", header=T) | |||
a<-levels(x[,8]) | |||
output = data.frame(i=character(), j=character(), cor = numeric(), p = numeric()); | |||
#k <-0; | |||
for(i in 1:22) { | |||
allele.i <- a[i]; | |||
vect.i <- x[which(x[,8]==allele.i),12]; | |||
for(j in (i+1):23) { | |||
allele.j <- a[j]; | |||
vect.j <-x[which(x[,8]==allele.j),12]; | |||
cor <- cor.test(vect.i,vect.j, method = "pearson"); | |||
output <- rbind (output, data.frame(i=allele.i, j=allele.j, cor=cor$estimate, p=cor$p.value)); | |||
} | |||
} | |||
write.table(output, "immune-output.txt", quote = F, sep = "\t") | |||
</syntaxhighlight> | |||
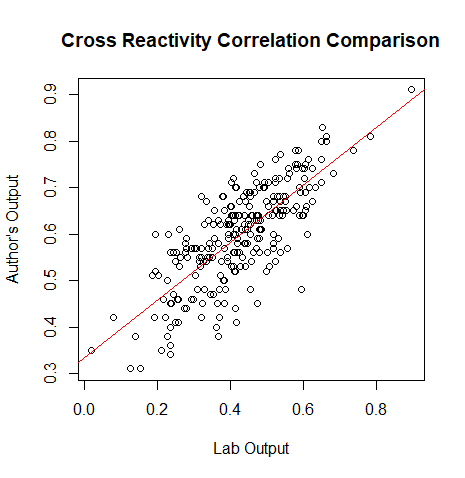
*(Muhammad) Creates a plot for the correlation values of the lab's data and the author's data | |||
<syntaxhighlight lang="bash"> | |||
#read in the authors cross reactivity correlation matrix | |||
cr<- read.csv("C:/ospc/matricesospc.csv", header=F, sep = ",") | |||
#puts all of the values of cr into corvect | |||
corvect<-c() | |||
for (i in 1:(nrow(cr)-1)) { | |||
for (j in (i+1):ncol(cr)) { | |||
corvect[length(corvect)+1]<- cr[i,j] | |||
} | |||
} | |||
#merging cross reactivity correlation data and the authors data | |||
df<- data.frame(output, corvect) | |||
#plots the dataframe | |||
plot(output[,3], corvect, main="Cross Reactivity Correlation Comparison", ylab = "Author's Output", xlab="Lab Output") | |||
#gives the liner model, relationship between our data and the authors | |||
b<-lm(corvect~output[,3]) | |||
#places the ab line on the plot | |||
abline(b, col=2) | |||
</syntaxhighlight> | |||
[[File:Rplot01.png|800px|thumb|left|Cross Reactivity Correlation Comparison]] | |||
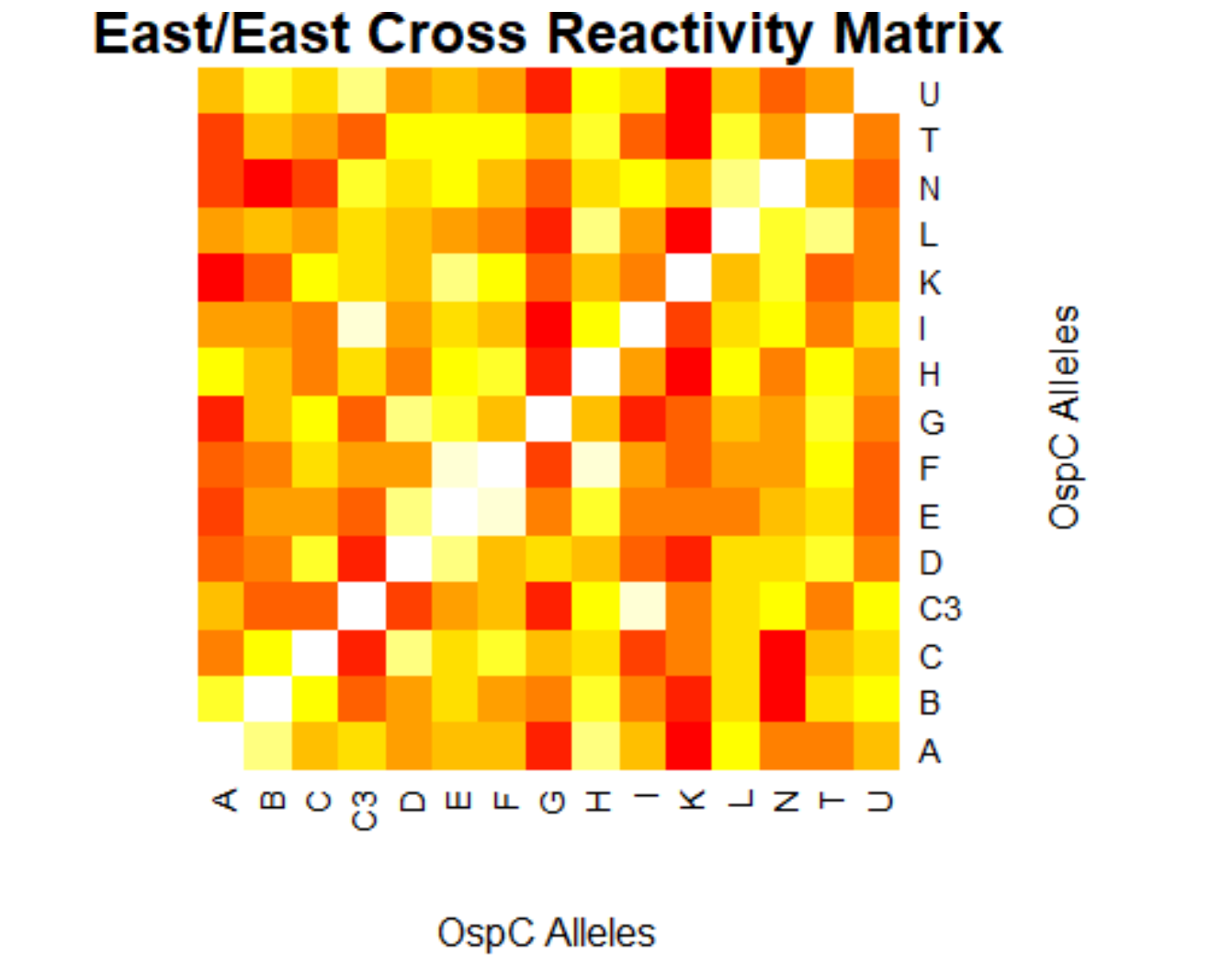
[[File:easteast.png|800px|thumb|left|East/East Cross Reactivity Matrix]] | |||
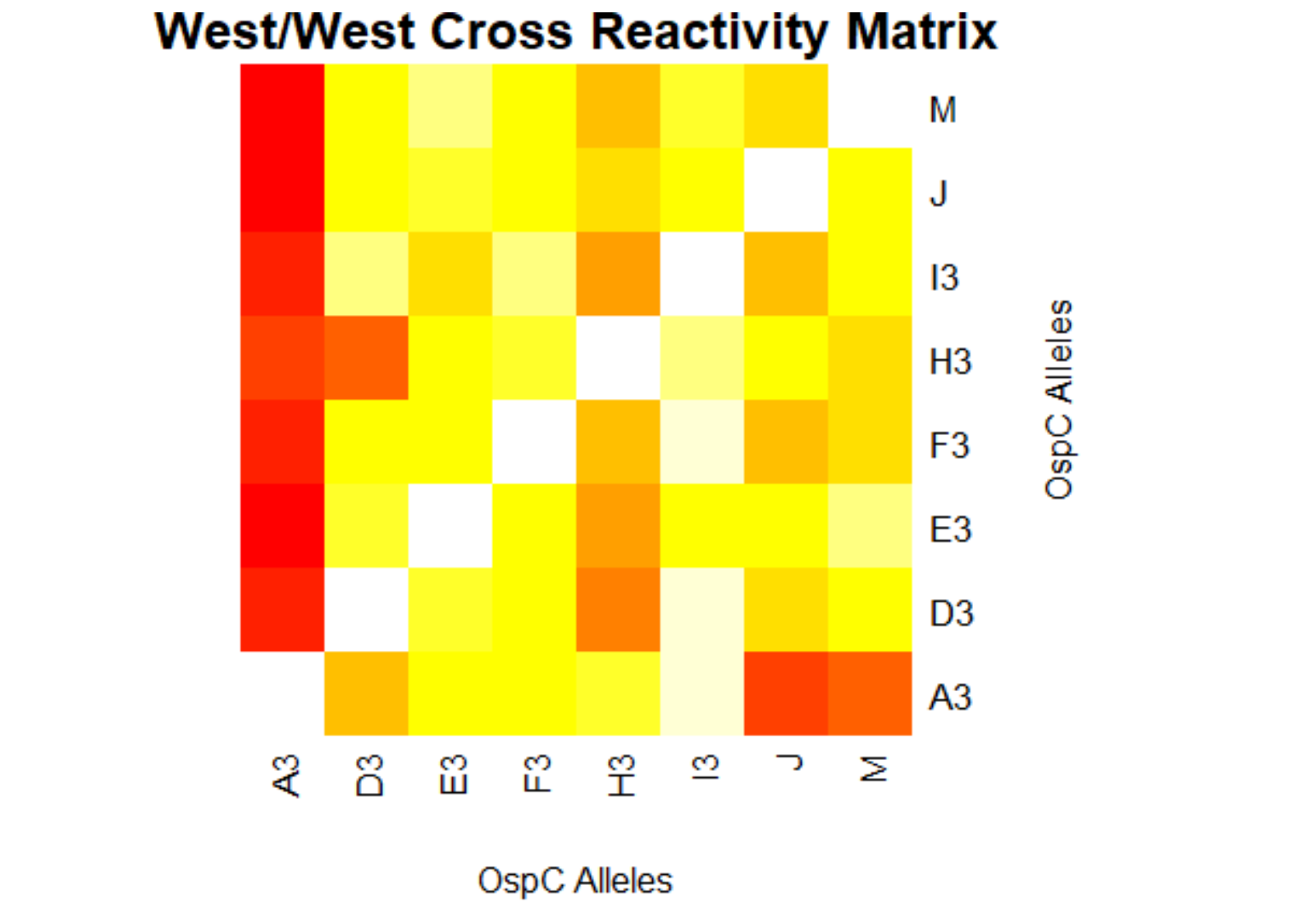
[[File:westwest.png|800px|thumb|left|West/West Cross Reactivity Matrix]] | |||
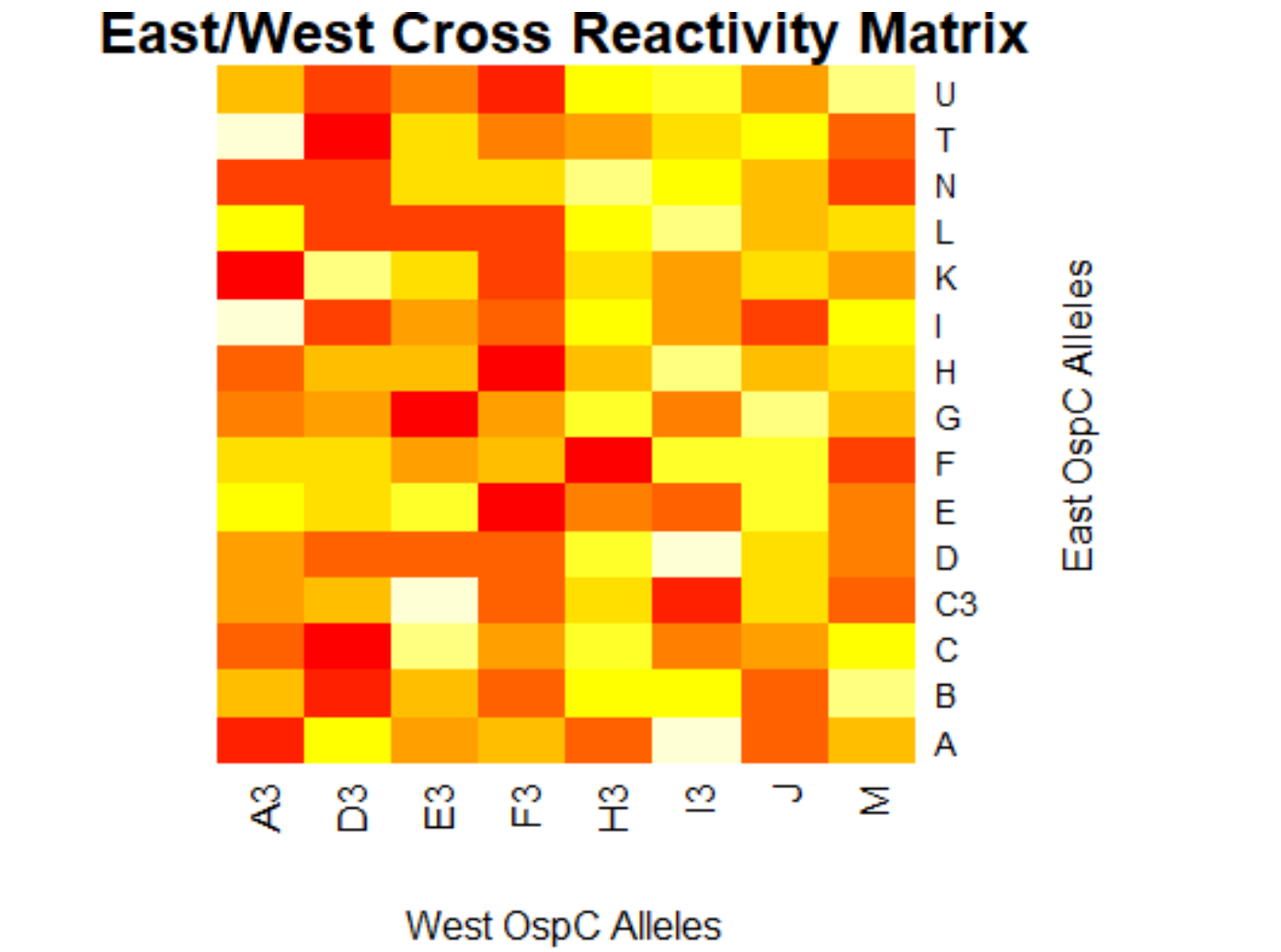
[[File:eastwest.png|800px|thumb|left|West/West Cross Reactivity Matrix]] | |||
Latest revision as of 21:28, 5 August 2018
Rules of Conduct
- No eating, drinking, or loud talking in the lab. Socialize in the lobby only.
- Be respectful to each other, regardless of level of study
- Be on time & responsible. Communicate in advance with the PI if late or absent
Participants
- Dr Oliver Attie, Research Associate
- Brian Sulkow, Research Associate
- Saymon Akther, CUNY Graduate Center, EEB Program
- Lily Li, CUNY Graduate Center, EEB Program
- Mei Wu, Bioinformatics Research Assistant
- Yinheng Li, Informatics Research Assistant
- Christopher Panlasigui, Hunter Biology
- Dr Lia Di, Senior Scientist
- Dr Weigang Qiu, Principal Investigator
- Summer Interns: Muhammad, Pavan, Roman, Benjamen, Andrew, Michelle, Hannah
Journal Club
- a Unix & Perl tutorial
- A short introduction to molecular phylogenetics: http://www.ncbi.nlm.nih.gov/pubmed/12801728
- A review on Borrelia genomics: https://www.ncbi.nlm.nih.gov/pubmed/24704760
- ospC epitope mapping: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0067445
- Codon usage changes fitness in E.coli: Frumkin et al (2018) "Codon usage of highly expressed genes affects proteome-wide translation efficiency". PNAS
Projects
Borrelia genome evolution (Led by Saymon)
- Goal 1. Estimate time of cross-Atlantic dispersal using core-genome sequences
- Goal 2. Investigate codon biases with respect to levels of gene expression. Data file:
* Andrew's BioPython code to calculate CAI
#Opens the fasta file and reads contents into a string (myStr).
myFile = open("B31-cp26.txt","r")
myStr = myFile.read()
myFile.close()
#Imports codon usage module.
from Bio.SeqUtils import CodonUsage as cu
#Takes myStr and processes it into a list of sequences (FastaList).
FastaList = myStr.split(">")
FastaList = FastaList[1:]
IDList = []
EnterList = []
##Separates FastaList into a list of sequence IDs (IDList) and a list of sequences (EnterList).
for seq in FastaList:
IDList += [seq[:6]]
EnterList += [seq[6:]]
##Removes enter characters from each sequence in EnterList.
SeqList = []
for seq in EnterList:
SeqStr = seq.replace("\n", "")
SeqList += [SeqStr]
#Calculates and presents the CAI value for each sequence using functions from the module.
myObject = cu.CodonAdaptationIndex()
myObject.generate_index("B31-cp26.txt")
for SeqIndex in range(len(SeqList)):
print (IDList[SeqIndex], ' CAI =', myObject.cai_for_gene(SeqList[SeqIndex]))
Output for cp26:
BB_B01 CAI = 0.7190039074113422
BB_B02 CAI = 0.678404951527374
BB_B03 CAI = 0.6893076488255271
BB_B04 CAI = 0.7250154635421513
BB_B05 CAI = 0.6971190458423587
BB_B06 CAI = 0.67042305582205
BB_B07 CAI = 0.6971020959083346
BB_B09 CAI = 0.6786931743972611
BB_B10 CAI = 0.7224886929887183
BB_B12 CAI = 0.6997502136447451
BB_B13 CAI = 0.7592966148479222
BB_B14 CAI = 0.6959525612884284
BB_B16 CAI = 0.6835709626613392
BB_B17 CAI = 0.6974779110749645
BB_B18 CAI = 0.7052250722958308
BB_B19 CAI = 0.7049049245887261
BB_B22 CAI = 0.6860641572293008
BB_B23 CAI = 0.6915165725213809
BB_B24 CAI = 0.7025276490965267
BB_B25 CAI = 0.7439914547011712
BB_B26 CAI = 0.7255623088410704
BB_B27 CAI = 0.7161378416520467
BB_B28 CAI = 0.7316661839512337
BB_B29 CAI = 0.6919705705489939
- Codon bias by Shannon information: BASH pipeline
- Simulate 100 sequences for each CDS:
cat filename.txt | while read line; do ./codon-info-sim.pl -n 100 BbB31.cutg.GCG "$line".fas > "$line"-sim.fas; done; - Calculate Shannon index for each CDS (simulated & actual)
./codon-info.pl BbB31.cutg.GCG foo.fas
- Simulate 100 sequences for each CDS:
Identification of host species from ticks (Led by Lily [after first-level])
- Goal 1. Protocol optimization for PCR amplification of host DNA from ticks
- Goal 2. Protocol development: library construction for MiSeq
- Goal 3. Development of bioinformatics protocols and sequence database
Pseudomonas Genome-wide Association Studies (GWAS) (Led by Mai & Yinheng, in collaboration with Dr Xavier of MSKCC)
- Goal 1. Association of genes/SNPs with biofilm formation and c-di-GMP levels: Manuscript preparation
- Goal 2. Association of genome diversity with metabolic diversity
- (Christopher) This script parses excel peak-area file into database & R inputs
#!/usr/bin/perl -w
use strict;
use Data::Dumper;
use Getopt::Std;
my %opts;
my $line_ct = 0;
my (@colnames, @areas, %seen_cmps, %seen_gids);
getopts('dr', \%opts);
while(<>) {
chomp;
$line_ct++;
next unless $line_ct >=4;
if ($line_ct == 4) {
@colnames = split "\t", $_;
for (my $i=5; $i<=$#colnames; $i++) { $seen_gids{$colnames[$i]}++ } # get uniq gids
next;
}
my @data = split "\t", $_;
$seen_cmps{$data[1]}++; # get unique compound formula
for (my $i=5; $i<=$#data; $i++) {
my $area = { 'compound' => $data[1], 'gid' =>$colnames[$i], 'peak_area' => $data[$i]};
push @areas, $area;
}
}
if ($opts{d}) { # for database output
foreach my $cmp (sort keys %seen_cmps) {
foreach my $gid (sort keys %seen_gids) {
my @peaks = grep { $_->{compound} eq $cmp && $_->{gid} == $gid } @areas;
my $peak_str = join ",", map {$_->{peak_area} || "NULL"} @peaks;
print join "\t", ($gid, $cmp, "{" . $peak_str . "}");
print "\n";
}
}
}
if ($opts{r}) { # for R output
foreach my $cmp (sort keys %seen_cmps) {
foreach my $gid (sort keys %seen_gids) {
my @peaks = grep { $_->{compound} eq $cmp && $_->{gid} == $gid } @areas;
foreach my $peak (@peaks) {
next unless $peak->{peak_area};
print join "\t", ($peak->{peak_area}, $gid, $cmp);
print "\n";
}
}
}
}
exit;
- (Benjamen) Creates a sample simple force directed network in R using networkD3. Install packages "igraph" and "networkD3".
library(igraph)
library(networkD3)
nodes <- c(rep('x1', 10), rep('x2',10), rep('x3',10), rep('x4',10))
ids <- c(1,1,1,-1,0,-1,0,0,-1,0,0,0,0,0,0,0,0,0,1,-1,0,0,0,0,0,1,-1,-1,0,0,0,0,0,1,-1,0,0,0,0,0)
df <- data.frame(nodes, ids)
df$ids
matrix(df$ids, nrow = 4, ncol = 10, byrow = T)
testmat<-matrix(df$ids, nrow = 4, ncol = 10, byrow = T)
rownames(testmat) <- c("x1", "x2", "x3", "x4")
colnames(testmat) <- c("v1", "v2", "v3", "v4", "v5", "v6", "v7", "v8", "v9", "v10")
testmat.inc <- graph_from_incidence_matrix(testmat)
testmat.edge <- as_edgelist(testmat.inc)
colnames(testmat.edge) <- c("From", "To")
testmat.df <- as.data.frame(testmat.edge)
simpleNetwork(testmat.df, opacity = .8, fontSize = 16)
- (Benjamen) Force-Directed Network Graph Visualization of the iJN746 S Matrix (Only Metabolites in Our Database)
Force-Directed Network Graph of iJN746 (Only Metabolites in Our Database)
library(igraph)
library(networkD3)
#create a preliminary edgelist from the original matrix. If using the simpleNetwork() function, this
#is the only required step.
stwo <- read.table("Stwo.tsv", header=T)
stwo.inc <- graph_from_incidence_matrix(stwo)
stwo.edge <- as_edgelist(stwo.inc)
colnames(stwo.edge) <- c("from", "to")
stwo.df <- as.data.frame(stwo.edge)
#both the "from" and "to" columns must be given a unique numerical value, as this is the only way
#forceNetwork() will run. To assign numerical values, we index each unique instance of a metabolite
#or reaction in order.
#index "from" column
unique(stwo.edge[,1])
order(unique(stwo.edge[,1]))
from.idx <- data.frame(row.names=unique(stwo.edge[,1]), row.idx=1:82)
stwo.df$from
from.idx[as.character(stwo.df$from),1]
stwo.df$from.idx <- from.idx[as.character(stwo.df$from),1]
#index "to" column
unique(stwo.edge[,2])
order(unique(stwo.edge[,2]))
to.idx <- data.frame(row.names=unique(stwo.edge[,2]), row.idx=1:188)
stwo.df$to
to.idx[as.character(stwo.df$to),1]
stwo.df$to.idx <- to.idx[as.character(stwo.df$to),1]
#add 81 to every value in "to" column and subtract 1 from every value in "from" column. This is so
#the indexing starts from 0, and every indexed variable on the "to" side continues from where "from"
#left off, which, in this case, is variable #82.
stwo.df$to.idx <- stwo.df[,4]+81
stwo.df$from.idx <- stwo.df[,3]-1
#create new data frame with just the "from" and "to" columns.
stwo.new <- data.frame(stwo.df$from.idx, stwo.df$to.idx)
colnames(stwo.new) <- c("from", "to")
#order the edgelist by the to column. The ordered "to" column is then appended to the "from" column,
#and the now disordered "from" column is appended to the "to" column, effectivley mirroring the original
#edgelist. This is done because every variable in the network must show up in the "from" column for
#the forceNetwork() function to work.
stwo.new2 <- stwo.new[order(stwo.new$to),]
#zoom in and out by rolling mouse wheel or double clicking. Click and drag to pan the model.
forceNetwork(Links = links, Nodes = nodes, Source="from", Target = "to", NodeID = "idn", Group = "type.label",
linkWidth = 1, linkColour = "green", fontSize = 20, zoom = T, legend = T, Nodesize = 6, opacity = 1,
charge = -50, width = 1920, height = 1080)
Machine learning approaches to evolution (Led by Oliver & Brian)
- Goal 1. Implement Hopfield network for optimization of protein structure
- Goal 2. Neural-net models of OspC. Structural alignment (S2 from Baum et al 2013):
- Goal 3. K-mer-based pipeline for genome classification
Weekly Schedule
- Summer kickoff (June 1, 2018, Friday): Introduction & schedules
- Week 1 (June 4-8):
- Monday: the Unix & Perl Tutorial, Part 1
- Tuesday: Unix Part2. Explore the "iris" data set using R, by following the the Monte Carlo Club Week 1 (1 & 2) and Week 2. Read McKay (2003), Chapters 38 & 39
- Thursday: 1st field day (Caumsett State Park); Participants: John, Muhammad, Pavan, Andrew, Dr Sun, Weigang, with 3 members of Moses team from Suffolk County Vector Control. Got ~110 deer tick nymphs
- Friday: meeting with MSKCC group at 11am; BBQ afterwards
- Week 2 (June 11-15):
- Monday: Lab meeting, projects assigned
- Tuesday: neural net tutorial (by Brian)
- Thursday: 2nd field day (Fire Island National Seashore). Participants: John, Brian, Mei, Muhammad, Pavan, Benjamin, and Weigang. Got ~100 lone-star ticks and 4 deer tick nymphs
- Week 3 (June 18-22):
- Monday: Lab meeting, 1st project reports
- Codon Bias: Theory, Coding, and Data (Andrew, Pavan, Saymon)
- OspC epitope identification: Serum correlation, sequence correlation, immunity-sequence correction (Muhammad, Roman, Brian)
- Pseudomonas metabolomics: parsing intensity file; theory & parsing SMBL file (Chris & Benjamin)
- Tuesday: working groups
- Wed: working groups
- Thursday: Big Data Workshop
- Friday: working groups
- Monday: Lab meeting, 1st project reports
- Week 4 (June 25-29):
- Monday: Lab meeting
- Tu-Th: work sessions
- Friday: Joint lab meeting with MSKCC/Lab visit
- Week 5 (July 2-6):
- Monday: Lab meeting
- Tuesday/Wed: 4th of July Break
- Thursday & Friday: work sessions
- Week 6 (July 9-13)
- Monday (July 9): E-reports & Wiki posts
- Week 7 (July 16-20)
- Monday (July 16): E-reports * Wiki posts
- Week 8 (July 23-27)
- Monday: lab meeting resumes
- Monte Carlo Club, Season 3 (Evolutionary Computing) starts
- Week 9 (July 30-Aug 3)
- Week 10 (Aug 6-Aug 10)
- Final report
Lab notes for Summer HS Interns
- NCI Cloud: Seven Bridges Cloud Platform. Create an user account
- Read documentation & tutorials: Documentation
Notes & Scripts
- (Weigang) Re-analysis of ospC cross-reactivity using normalization:
- Fold change vs alleles for mouse + humans: interactive heatmap by heatmaply
- Correlation matrix based on N=23 mouse samples (and 7 controls): interactive heatmap by heatmaply
- Correlation matrix based on N=55 human samples (and 25 controls): interactive heatmap by heatmaply
- Correlation matrix based on both mouse & human samples: interactive heatmap by heatmaply]
- (Weigang) A sample R script to parse Table S2 from Baum et al 2013, sera-antigen reactivity measurements
# preliminaries: save as TSV; substitute "\r" if necessary;
# substitute "N/A" to "NA"; remove extra columns
setwd("Downloads/")
x <- read.table("table-s2.txt4", sep="\t", header=T)
View(x)
colnames(x)
which(x[,8]=="A")
x[which(x[,8]=="A"),12]
x[which(x[,8]=="A3"),12]
cor.test(x[which(x[,8]=="A3"),12], x[which(x[,8]=="A"),12], method = "pearson")
x.cor$estimate
levels(x[,8]) # obtain ospC allele types; to be looped through in pairwise
for (i in 1:?) { for (j in ?:?) {cor.test(....)}}
- (Muhammad) Output generates data frame of correlation/p values for 23 different Osp-C allele types in pairwise
setwd("C:/R_OspC")
x <- read.table("Table-S2.txt", sep="\t", header=T)
a<-levels(x[,8])
output = data.frame(i=character(), j=character(), cor = numeric(), p = numeric());
#k <-0;
for(i in 1:22) {
allele.i <- a[i];
vect.i <- x[which(x[,8]==allele.i),12];
for(j in (i+1):23) {
allele.j <- a[j];
vect.j <-x[which(x[,8]==allele.j),12];
cor <- cor.test(vect.i,vect.j, method = "pearson");
output <- rbind (output, data.frame(i=allele.i, j=allele.j, cor=cor$estimate, p=cor$p.value));
}
}
write.table(output, "immune-output.txt", quote = F, sep = "\t")
- (Muhammad) Creates a plot for the correlation values of the lab's data and the author's data
#read in the authors cross reactivity correlation matrix
cr<- read.csv("C:/ospc/matricesospc.csv", header=F, sep = ",")
#puts all of the values of cr into corvect
corvect<-c()
for (i in 1:(nrow(cr)-1)) {
for (j in (i+1):ncol(cr)) {
corvect[length(corvect)+1]<- cr[i,j]
}
}
#merging cross reactivity correlation data and the authors data
df<- data.frame(output, corvect)
#plots the dataframe
plot(output[,3], corvect, main="Cross Reactivity Correlation Comparison", ylab = "Author's Output", xlab="Lab Output")
#gives the liner model, relationship between our data and the authors
b<-lm(corvect~output[,3])
#places the ab line on the plot
abline(b, col=2)