Annotate-a-genome: Difference between revisions
Jump to navigation
Jump to search
imported>Weigang |
imported>Weigang |
||
| Line 85: | Line 85: | ||
* [http://www.ncbi.nlm.nih.gov/nuccore/CP006647.1 main chromosome] | * [http://www.ncbi.nlm.nih.gov/nuccore/CP006647.1 main chromosome] | ||
|| Accession: CP006647; Assigned to: LLW | || Accession: CP006647; Assigned to: LLW | ||
|- | |||
| 107 || 94a || B. burgdorferi (Northeast US) || Lyme Disease || | |||
* [http://www.ncbi.nlm.nih.gov/nuccore/ABGK02000008.1 main chromosome] | |||
|| Accession: ABGK02000008; Assigned to: QZ | |||
|} | |} | ||
Revision as of 02:55, 9 March 2014
Project Goals
- Annotate and add newly sequenced Borrelia genomes to BorreliaBase
- Build an informatics pipeline for gene prediction, ortholog calls, databasing, and synteny analysis
Download genome sequences from GenBank
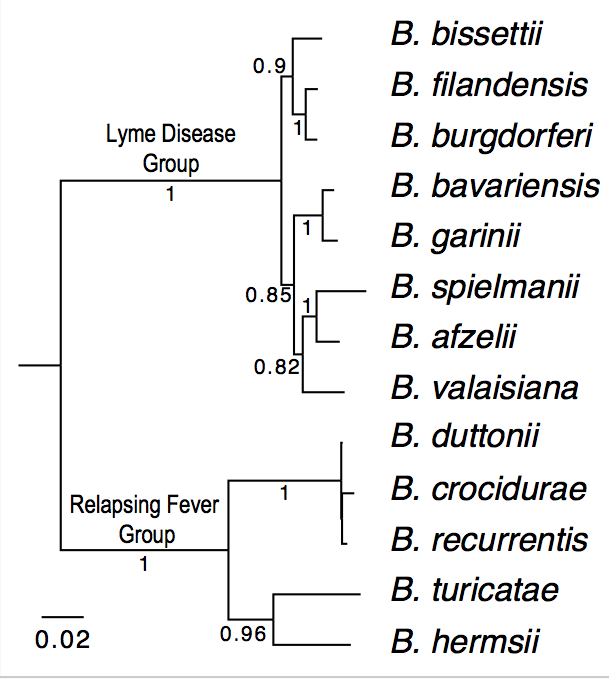
| Genome_id | Strain | Species | Group | Genome Sequences | Notes |
|---|---|---|---|---|---|
| 100 | B31 | B. burgdorferi (reference genome) | Lyme Disease | Reference. Already downloaded as "ref.pep" | |
| 114 | CA382 | B. burgdorferi (California) | Lyme Disease | Accession: CP005925; Assigned to: HA | |
| 115 | CA8 | B. burgdorferi (California) | Lyme Disease |
| |
| 304 | BgVir | B. garinii (Russia) | Lyme Disease |
| |
| 305 | NMJW1 | B. garinii (China) | Lyme Disease | Accession: CP003866.1; Assigned to: AL | |
| 402 | HLJ01 | B. afzelii (China) | Lyme Disease | Accession: CP003882; Assigned to: RL | |
| 1003 | Ly | B. duttonii (Tanzania) | Relapsing Fever |
|
|
| 1001 | A1 | B. recurrentis (Ethiopia) | Relapsing Fever |
| |
| 1100 | DAH | B. hermsii (Washington State) | Relapsing Fever | Accession: CP000048; Assigned to: KR | |
| 1200 | 91E135 | B. turicatae (Texas) | Relapsing Fever | Accession: CP000049; Assigned to: MDR | |
| 1002 | Achema | B. crocidurae (Mauritania) | Relapsing Fever |
|
Accession: CP003426; Assigned to: VS |
| 1400 | HR1 | B. parkeri (??) | Relapsing Fever | Accession: CP0007022; Assigned to: AV | |
| 1300 | LB-2001 | B. miyamotoi (Northeast US) | Relapsing Fever | Accession: CP006647; Assigned to: LLW | |
| 107 | 94a | B. burgdorferi (Northeast US) | Lyme Disease | Accession: ABGK02000008; Assigned to: QZ |
Protocol
Dependencies
- BASH (default shell of Linux OS and Apple OS X)
- Perl and BioPerl
- DNATweezer
- NCBI Standalone BLAST+
Fetch genome sequences
- Commands:
./bioseq -z 'b31_accession' -o 'genbank' > b31.gb # Reference genome for ortholog identification. Choose main, cp26, or lp54
./bioseq -z 'gb_accession' -o 'genbank' > new.gb
./gb2fas -n b31.gb > b31.nuc # Extract CDS
./gb2fas -n new.gb > new.nuc
./bioseq -t b31.nuc > b31.pep # Translate (and remove those with internal stop codons)
./bioseq -t new.nuc > new.pep
- Perl code for "gb2fas.pl":
#!/usr/bin/env perl
# Extract sequences from a GenBank file
# Input: a GenBank file
# Output: -n: CDS sequences in FASTA; -t: CDS information in Tab-delimited
use strict;
use Bio::SeqIO;
use Getopt::Std;
use Data::Dumper;
use 5.10.0;
my %opts;
getopts('tn',\%opts);
die "$0 [-nt] <genbank_file>\n" unless @ARGV == 1;
my $gb_file = shift @ARGV;
my $in = Bio::SeqIO->new(-file=>$gb_file, -format=>'genbank');
my $cds_ct=0;
while (my $seqobj = $in->next_seq() ) {
my @features = $seqobj->get_SeqFeatures(); # just top level
foreach my $feat ( @features ) {
next unless $feat->primary_tag eq "CDS";
$cds_ct++;
&to_db($feat, $cds_ct) if $opts{t};
&to_nt($feat, $seqobj) if $opts{n};
}
}
exit;
sub to_nt {
my $ft = shift;
my $seq = shift;
say ">", $ft->get_tag_values("locus_tag");
my $subseq = $seq->trunc($ft->start, $ft->end);
if ($ft->strand > 0) {
say $subseq->seq();
} else {
say $subseq->revcom()->seq();
}
}
sub to_db {
my $ft = shift;
my $ct = shift;
my $orf_id = sprintf "ORF%04d", $ct;
my $gid = 401; # this is bad and needs improvement: nothing should be hard-coded
my $con_id = 111114823; # the same problem
my $locus = sprintf "%s", $ft->get_tag_values('locus_tag');
my $prod = sprintf "%s", $ft->get_tag_values('product');
$prod =~ tr/\'/_/;
my $strand = ($ft->strand > 0) ? 't' : 'f';
say join "\t", ($gid, $con_id, $orf_id, 'f', $ft->start, $ft->end, $strand, $locus, $prod);
}
Predict orthologs with reciprocal BLAST
- Commands:
makeblastdb -in b31.pep -parse_seqids # Prepare the reference DB
makeblastdb -in new.pep -parse_seqids # Prepare the new genome DB
blastp -query new.pep -db b31.pep -outfmt 6 -evalue 1e-3 -out forward_blast.out # Forward BLAST
blastp -query b31.pep -db new.pep -outfmt 6 -evalue 1e-3 -out reverse_blast.out # Reverse BLAST
./check-reciprocal.pl forward_blast.out reverse_blast.out > new.orthlogs 2> new.not-orthologs # Identify orthologs
- Code for "check-reciprocal.pl":
#!/usr/bin/env perl
use strict;
use warnings;
use Data::Dumper;
die "$0 <forward_blast_output> <reverse_blast_output>\n" unless @ARGV == 2;
my ($fwd, $rev) = @ARGV;
my (@fwd_top_hits, @rev_top_hits);
open FWD, "<" . $fwd;
my (%fwd_top_hits, %rev_top_hits, @query);
my $query_ct=0;
while (<FWD>) {
chomp;
my @data = split;
next if $fwd_top_hits{$data[0]};
$fwd_top_hits{$data[0]} = $data[1];
push @query, $data[0];
$query_ct++;
}
close FWD;
warn "Total query having hits:" . $query_ct . "\n";
open REV, "<" . $rev;
while (<REV>) {
chomp;
my @data = split;
next if $rev_top_hits{$data[0]};
$rev_top_hits{$data[0]} = $data[1];
}
close REV;
foreach my $q (@query) { # e.g., BafPKo_0002
my $top = $fwd_top_hits{$q}; # e.g. BB_0002
if ( $q eq $rev_top_hits{$top}) {
print "Found reciprocol top hits:\t", $q, "\t", $top, "\n";
} else {
warn "Not reciprocol top hits:\t", $q, "\t", $top, "\t", $rev_top_hits{$top}, "\n";
}
}
exit;
Verify with synteny broswer
./gb2fas -t new.gb > new-to-orf-table.txt
# load into database with SQL
# Visualize synteny