QuBi/bio203: Difference between revisions
imported>Weigang mNo edit summary |
imported>Weigang mNo edit summary |
||
| Line 20: | Line 20: | ||
## NCBI GeneID | ## NCBI GeneID | ||
## Chromosome location | ## Chromosome location | ||
## | ## Click on "GenBank" and identify its gene structure, including the length of primary transcript, coding sequences, 5'-UTR and 3'-UTR. Does it have any introns? | ||
## Zoom out to find its neighboring genes. Zoom out to read DNA sequences. | ## Zoom out the Sequence View to find its neighboring genes. Zoom out to read DNA sequences. | ||
# Click the link to OMIM (under '''Phenotype''') and find phenotypes associated with TAS2R38 gene | # Click the link to OMIM (under '''Phenotype''') and find phenotypes associated with TAS2R38 gene | ||
## What does OMIM stand for? | ## What does OMIM stand for? | ||
| Line 28: | Line 28: | ||
## Is the correlation between TAS2R38 gene variations and the PTC phenotype variations 100%? If not, what could be the other causes? | ## Is the correlation between TAS2R38 gene variations and the PTC phenotype variations 100%? If not, what could be the other causes? | ||
== | ==Part 2. Predict results of restriction analysis== | ||
This sequence | This gene sequence of TAS2R38 is pasted in the following: | ||
<div style="font-family:Monospace;line-height:1;width:400px;border-style:solid;border-width:1px;border-color:#AAAAFF;background-color:#EEEEFF;padding-left:5px;padding-right:5px;padding-top:0px;padding-bottom:10px;"> | <div style="font-family:Monospace;line-height:1;width:400px;border-style:solid;border-width:1px;border-color:#AAAAFF;background-color:#EEEEFF;padding-left:5px;padding-right:5px;padding-top:0px;padding-bottom:10px;"> | ||
CAGTCTTCCACCCTATGATAAGCTCTTACGTGTATCCAAGAGATGTTCTAGAGAAACAACATCCCTCTAA | |||
GTTTCCTGCCAGAACTTTTTATGCGCTCGCTTTGGGATAGATCTAGGCAAAGAGCTGGATGCTTTGTGAA | |||
GGAAAGGTCCTGGCTTGGAACGTACATTTACCTTTCTGCACTGGGTGGCAACCAGGTCTTTAGATTAGCC | |||
AACTAGAGAAGAGAAGTAGAATAGCCAATTAGAGAAGTGACATCATGTTGACTCTAACTCGCATCCGCAC | |||
TGTGTCCTATGAAGTCAGGAGTACATTTCTGTTCATTTCAGTCCTGGAGTTTGCAGTGGGGTTTCTGACC | |||
AATGCCTTCGTTTTCTTGGTGAATTTTTGGGATGTAGTGAAGAGGCAGGCACTGAGCAACAGTGATTGTG | |||
TGCTGCTGTGTCTCAGCATCAGCCGGCTTTTCCTGCATGGACTGCTGTTCCTGAGTGCTATCCAGCTTAC | |||
CCACTTCCAGAAGTTGAGTGAACCACTGAACCACAGCTACCAAGCCATCATCATGCTATGGATGATTGCA | |||
AACCAAGCCAACCTCTGGCTTGCTGCCTGCCTCAGCCTGCTTTACTGCTCCAAGCTCATCCGTTTCTCTC | |||
ACACCTTCCTGATCTGCTTGGCAAGCTGGGTCTCCAGGAAGATCTCCCAGATGCTCCTGGGTATTATTCT | |||
TTGCTCCTGCATCTGCACTGTCCTCTGTGTTTGGTGCTTTTTTAGCAGACCTCACTTCACAGTCACAACT | |||
GTGCTATTCATGAATAACAATACAAGGCTCAACTGGCAGATTAAAGATCTCAATTTATTTTATTCCTTTC | |||
TCTTCTGCTATCTGTGGTCTGTGCCTCCTTTCCTATTGTTTCTGGTTTCTTCTGGGATGCTGACTGTCTC | |||
CCTGGGAAGGCACATGAGGACAATGAAGGTCTATACCAGAAACTCTCGTGACCCCAGCCTGGAGGCCCAC | |||
ATTAAAGCCCTCAAGTCTCTTGTCTCCTTTTTCTGCTTCTTTGTGATATCATCCTGTGCTGCCTTCATCT | |||
CTGTGCCCCTACTGATTCTGTGGCGCGACAAAATAGGGGTGATGGTTTGTGTTGGGATAATGGCAGCTTG | |||
TCCCTCTGGGCATGCAGCCATCCTGATCTCAGGCAATGCCAAGTTGAGGAGAGCTGTGATGACCATTCTG | |||
CTCTGGGCTCAGAGCAGCCTGAAGGTAAGAGCCGACCACAAGGCAGATTCCCGGACACTGTGCTGAGAAT | |||
GGACATGAAATGAGCTCTTCATTAATACGCCTGTGAGTCTTCATAAATATGCCTCTGATTCTTCAGGAAT | |||
ACAACTCTGATTCCTCACAAAGCCTTCCAATTTCTTCTATAAAACACAATTGAAAGTCTCTCCACTTTGT | |||
ATCAATGAACTCACTTATAGATGAATAAAATAATTAAGCACTATACATGGCCTAGGCAAGAATAATGTTG | |||
GTACCCTAGGTTTGT | |||
</div> | </div> | ||
# Identify | # Identify 5'-UTR, 3'-UTR, start codon, and stop codon. | ||
# | # Identify the regions your primers bind | ||
# | # Identify the base location that contains 785 C/T SNP | ||
===Identify regulatory elements through literature search=== | ===Identify regulatory elements through literature search=== | ||
Revision as of 07:04, 21 September 2013
- BIOL 203 Lab 4. Bioinformatics Exercises
Research in modern molecular genetics increasingly rely on genomic information and computation. The following exercises will expose you to the field of bioinformatics, including the use of online databases and statistical analysis of genetic data.
Introduction
DNA and its organization into genes makes up an organism's genotype. The expression and presentation of those genes in the organism's development, physiology, and physical appearance (physical traits) make up the phenotype of the organism. Phenotypic variations among individuals of a species (e.g., humans) are caused by genotype variations, environmental factors, and interactions between genetic and environmental factors. In other words, phenotypic variations among individuals often have complex, unclear mechanisms and are not necessarily due entirely to genetic differences.
In this lab section, we will explore the concepts of phenotype and genotype by looking at the variations in the TAS2R38 gene, which is responsible for part of the sensation of taste. The taste receptor protein TAS2R38 (taste receptor 2, member 38) has been associated with the ability to taste the bitter compound phenylthiocarbamide (PTC) (Kim et al. 2003). Although most people can taste PTC ("tasters"), a centain percentage of people cannot ("nontasters"). In this experiment, you will test your Taster phenotype as well as determine your Taster genotype and correlate the phenotype with the genotype data. Your results and those of your classmates will be combined to statistically validate if there is such a phenotype-genotype association.
Learning goals and outcomes
- Understand phenotype, genotype, and their association
- Be able to use the NCBI online databases
- Be able to predict genotype frequencies using Hardy-Weinberg equilibrium
- Be able to use the contingency test of genotype-phenotype associations
Part 1. Search for TAS2R38 gene information using NCBI online databases
- Point your browser to the NCBI Human Genome Resource page
- Type in the "Find A Gene" search box "TAS2R38" and select "Homo sapiens" from the pull-down menu. Click "Go"
- Select the first link, which leads to an NCBI Gene Card page. Use the Gene Card to identify the following information on TAS2R38 gene:
- NCBI GeneID
- Chromosome location
- Click on "GenBank" and identify its gene structure, including the length of primary transcript, coding sequences, 5'-UTR and 3'-UTR. Does it have any introns?
- Zoom out the Sequence View to find its neighboring genes. Zoom out to read DNA sequences.
- Click the link to OMIM (under Phenotype) and find phenotypes associated with TAS2R38 gene
- What does OMIM stand for?
- What are the expected "taster" and "nontaster" frequencies within human populations?
- If the ability to taste bitterness is evolutionary advantageous, how are alleles contributing to "nontaster" maintained in population?
- Is the correlation between TAS2R38 gene variations and the PTC phenotype variations 100%? If not, what could be the other causes?
Part 2. Predict results of restriction analysis
This gene sequence of TAS2R38 is pasted in the following:
CAGTCTTCCACCCTATGATAAGCTCTTACGTGTATCCAAGAGATGTTCTAGAGAAACAACATCCCTCTAA GTTTCCTGCCAGAACTTTTTATGCGCTCGCTTTGGGATAGATCTAGGCAAAGAGCTGGATGCTTTGTGAA GGAAAGGTCCTGGCTTGGAACGTACATTTACCTTTCTGCACTGGGTGGCAACCAGGTCTTTAGATTAGCC AACTAGAGAAGAGAAGTAGAATAGCCAATTAGAGAAGTGACATCATGTTGACTCTAACTCGCATCCGCAC TGTGTCCTATGAAGTCAGGAGTACATTTCTGTTCATTTCAGTCCTGGAGTTTGCAGTGGGGTTTCTGACC AATGCCTTCGTTTTCTTGGTGAATTTTTGGGATGTAGTGAAGAGGCAGGCACTGAGCAACAGTGATTGTG TGCTGCTGTGTCTCAGCATCAGCCGGCTTTTCCTGCATGGACTGCTGTTCCTGAGTGCTATCCAGCTTAC CCACTTCCAGAAGTTGAGTGAACCACTGAACCACAGCTACCAAGCCATCATCATGCTATGGATGATTGCA AACCAAGCCAACCTCTGGCTTGCTGCCTGCCTCAGCCTGCTTTACTGCTCCAAGCTCATCCGTTTCTCTC ACACCTTCCTGATCTGCTTGGCAAGCTGGGTCTCCAGGAAGATCTCCCAGATGCTCCTGGGTATTATTCT TTGCTCCTGCATCTGCACTGTCCTCTGTGTTTGGTGCTTTTTTAGCAGACCTCACTTCACAGTCACAACT GTGCTATTCATGAATAACAATACAAGGCTCAACTGGCAGATTAAAGATCTCAATTTATTTTATTCCTTTC TCTTCTGCTATCTGTGGTCTGTGCCTCCTTTCCTATTGTTTCTGGTTTCTTCTGGGATGCTGACTGTCTC CCTGGGAAGGCACATGAGGACAATGAAGGTCTATACCAGAAACTCTCGTGACCCCAGCCTGGAGGCCCAC ATTAAAGCCCTCAAGTCTCTTGTCTCCTTTTTCTGCTTCTTTGTGATATCATCCTGTGCTGCCTTCATCT CTGTGCCCCTACTGATTCTGTGGCGCGACAAAATAGGGGTGATGGTTTGTGTTGGGATAATGGCAGCTTG TCCCTCTGGGCATGCAGCCATCCTGATCTCAGGCAATGCCAAGTTGAGGAGAGCTGTGATGACCATTCTG CTCTGGGCTCAGAGCAGCCTGAAGGTAAGAGCCGACCACAAGGCAGATTCCCGGACACTGTGCTGAGAAT GGACATGAAATGAGCTCTTCATTAATACGCCTGTGAGTCTTCATAAATATGCCTCTGATTCTTCAGGAAT ACAACTCTGATTCCTCACAAAGCCTTCCAATTTCTTCTATAAAACACAATTGAAAGTCTCTCCACTTTGT ATCAATGAACTCACTTATAGATGAATAAAATAATTAAGCACTATACATGGCCTAGGCAAGAATAATGTTG GTACCCTAGGTTTGT
- Identify 5'-UTR, 3'-UTR, start codon, and stop codon.
- Identify the regions your primers bind
- Identify the base location that contains 785 C/T SNP
Identify regulatory elements through literature search
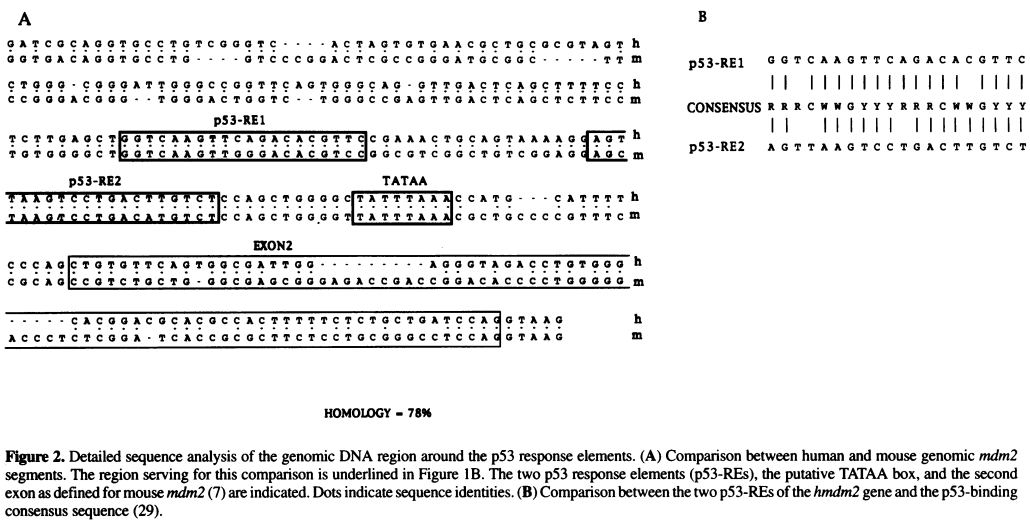
Source: Zauberman et al. "A functional p53-responsive intronic promoter is contained within the human mdm2 gene". Nucleic Acids Res. 1995 July 25; 23(14): 2584–2592. Pubmed PDF
- Identify the TATA box in the above sequence by looking for AT-rich regions
- Once the TATA box is located, use this sequence as the anchor point to locate other elements, including the two p53 Response Element (p53 RE) sites and the Exon 2
- Locate the EcoRI restriction site using the NEBcutter website
- What are the expected EcoRI fragment sizes for the two possible orientations of your cloned DNA?
- Which orientation would you expect to give higher luciferase expression?
Part 2. Perform statistical test of association between genotype and phenotype
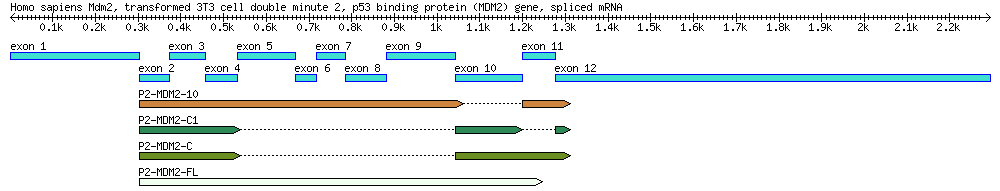
A diagram of the MDM2 gene used in this exercise, along with its splice variants. By the end of this module you will create a similar diagram. Reference: Arva NC, Talbott KE, Okoro DR, Brekman A, Qiu WG, Bargonetti J. 2008. Disruption of the p53-Mdm2 complex by Nutlin-3 reveals different cancer cell phenotypes. Ethnicity and Disease. 18(S2):1-8.
Key Concepts
- Use BLAST (not Google) to find matches of DNA and protein sequences
- Alternative splicing and isoforms of a single gene
You will use the following table for your exercise:
| Genbank Accession # | cDNA Clone | Description | Cell Line | Length (bp) |
|---|---|---|---|---|
| AF527840 | Genomic DNA | 34,088 | ||
| EU076746 | P2-MDM2-C1 | cDNA | MANCA | 427 |
| EU076747 | P2-MDM2-10 | cDNA | ML-1 | 842 |
| EU076748 | P2-MDM2-C | cDNA | A876 | 505 |
| EU076749 | P2-MDM2-FL | cDNA | SJSA-1 | 845 |
Explore the GenBank file
- Search GenBank using the accession AF527840. Read the GenBank file and find out from the feature table how many introns and exons this sequence has according to the "mRNA" and "CDS" features.
- DRAW a diagram of this gene using the information and coordinates listed in the annotation.
- Label the top of the diagram with basic information, such as the gene's name and species information.
- Label coordinates for introns, exons, 3'/5' UTRs,start-codon, and stop-codon coordinates.
- Draw the diagram mostly to scale. It does NOT have to be perfect, but make a reasonable effort. Put a scale bar and length markers on your drawing.
Explore the graphical presentation of the gene
Genbank provides graphical representations of the sequences on its database: click the "Graphics" link below the sequence title, OR click "Display Settings" above the title, and choose "Graphics". Take a few minutes to explore this graphical and compare it with your diagram.
Use BLAST to determine which exons are used in the mRNA transcript
This is the most "bioinformatic" part of the assignment. Blast one of the mRNA sequences (EU076746, EU076747, EU076748, EU076749) against the main sequence (AF527840) and use the results to answer the following questions. Suggested procedures:
- Go to the NCBI BLAST website
- Click the link “Align two (or more) sequences using BLAST (bl2seq)” under “Specialized BLAST” (near the page bottom)
- In the “Sequence 1” text box, type “AF527840” (the accession for the genomics). Fill in “from 1” and “to 34088”. In the “Sequence 2” text box, type in "EU076748" (or other cDNA accession in the table). Fill in the “from” box with 1 and the “to” box with 505.
- Click “Align”. You should get a “Blast Result” output page.
- Interpret your results:
- Which exons are present and which ones are absent in EU076746, EU076747, EU076748, EU076749? (Hint: Refer to the mRNA join statement).
- Explain the following BLAST terms: “Expect” (e-value), “Identities”, “Gap”, “Strand”