Spombe: Difference between revisions
imported>Lia |
imported>Weigang |
||
| Line 1: | Line 1: | ||
=Friday, June 13, 2014= | =Friday, June 13, 2014= | ||
Fold Change vs. Chrom Location Plots (batch 2) | Fold Change vs. Chrom Location Plots (batch 2) | ||
* [http:// | * [http://diverge.hunter.cuny.edu/spombe/fc-chrm1.html Chromosome 1] | ||
* [http:// | * [http://diverge.hunter.cuny.edu/spombe/fc-chrm2.html Chromosome 2] | ||
* [http:// | * [http://diverge.hunter.cuny.edu/spombe/fc-chrm3.html Chromosome 3] | ||
<br> | <br> | ||
Revision as of 13:30, 14 December 2018
Friday, June 13, 2014
Fold Change vs. Chrom Location Plots (batch 2)
Wednesday, March 5, 2014
MA Plots (batch 2)
Excel file:
Sunday, January 5, 2014
Interactive MA Plots (with Fc added in tooltips)
Monday, November 25, 2013
Pathway analysis (fold_change >=1)
- Molecular Function (s4 vs. s1):
- Biological Process (s4 vs. s1):
Wednesday, November 13-14, 2013
- Differentially expressed genes (fold_change >=1)
| Interactive MA Plots | Excel file |
|---|---|
| s4 vs. s1 | |
| s7 vs. s1 |
- Expression levels in cytosol & nuclei (fold_change of c/n ratio >= 1)
| Interactive Plots | Excel file |
|---|---|
| Strain 10 vs. 558 (78 genes) | |
| Strain 13 vs. 558 (83 genes) | |
| Strains 10, 13, and 558 (78+83-30=131 genes) |
Tuesday, November 12, 2013
Interactive MA Plots
Tuesday, November 5, 2013
Differentially expressed genes (fold_change >=2)
- s4 vs. s1, s7 vs.s1:
Monday, November 4, 2013
Linked plots for C/N ratio:
- Over-expression experiment:
- Knock-down experiment:
Sunday, November 3, 2013
Interactive MA Plots for C/N ratios
- Strain s5/s6 vs. s2/s3:
- Strain s8/s9 vs. s2/s3:
Saturday, November 2, 2013
Interactive MA Plots for total expression levels
- Strain s4 vs. s1:
- Strain s7 vs. s1:
Tuesday, October 30, 2013
Gene export affected by OE or KD of nup211: Dotplot of C/N ratios of genes expression
| Title | Plot | Excel file |
|---|---|---|
| Strain s4 vs. s1 | ||
| Strain s7 vs. s1 |
Monday, October 29, 2013
Differentially expressed genes (fold_change >=2)
| Title | Plot | Excel file |
|---|---|---|
| MAPlot of 158 genes (s4 vs. s1) | ||
| MAPlot of 240 genes (s7 vs. s1) |
Friday, October 25, 2013
Comparison of 3 Samples with total mRNA (We debugged rows with multiple gene IDs)
| Title | Plot | Plot (scaled by gene) | Excel file |
|---|---|---|---|
| Heatmap of 90 significant genes in 3 samples |
Wednesday, October 23, 2013
Comparison of 3 Samples with total mRNA
| Title | Plot | Plot (scaled by gene) | Excel file |
|---|---|---|---|
| Heatmap of 89 significant genes in 3 samples |
Thursday, October 17, 2013
mRNA export affected by KD or OE of nup211
| Title | Plot |
|---|---|
| Export (C/N) of 40 genes affected by KD of nup211 (heatmap) | |
| Export (C/N) of 39 genes affected by OE of nup211 (heatmap) |
Wednesday, October 16, 2013
Comparison of 4 Samples with total mRNA
| Title | Plot | Plot (scaled by gene) |
|---|---|---|
| Heatmap of 76 significant genes |
Friday, October 11, 2013
Comparison of all 10 Samples
| Title | Plot | Plot (scaled by gene) |
|---|---|---|
| Barplot Nup 211 | ||
| Heatmap of 13 genes of interest | ||
| Heatmap of 80 significant genes | ||
| Heatmap of 39 genes of interest |
Tuesday, October 8, 2013
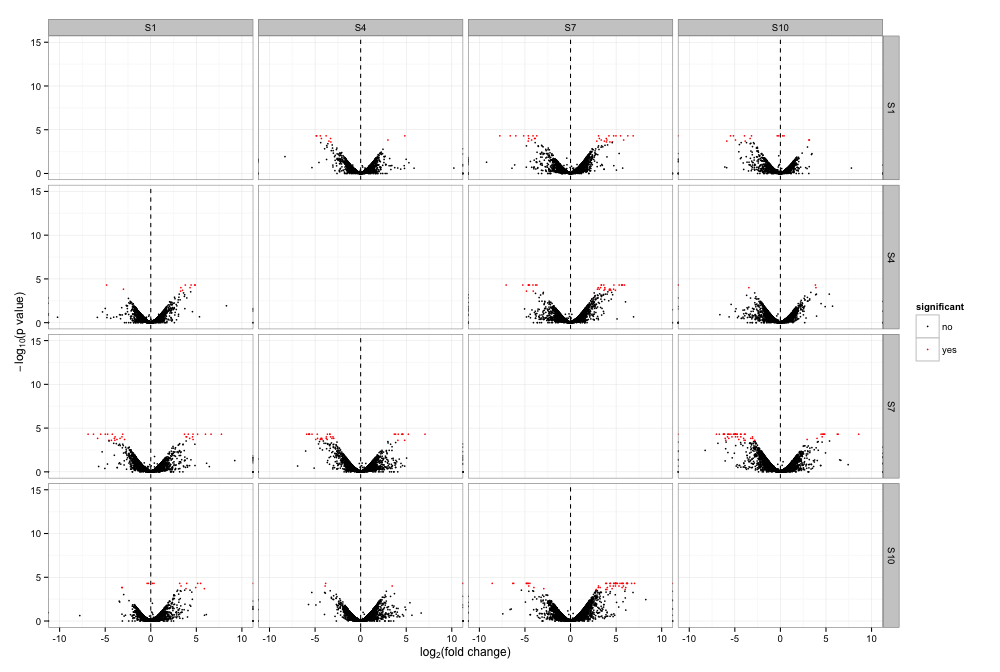
Comparison between Samples 1,4,7, and 10
| Title | Plots |
|---|---|
| Volcano Plots | |
| Heatmap of 13 genes of interest | |
| Heatmap of 76 significant genes |
References
- Trapnell et al (2012). Nature Protocol. 7(2): 562.
- Goff et al (2012). CummeRbund Manual