Summer 2018: Difference between revisions
Jump to navigation
Jump to search
imported>Weigang |
imported>Weigang |
||
| Line 34: | Line 34: | ||
# Goal 2. Association of genome diversity with metabolic diversity | # Goal 2. Association of genome diversity with metabolic diversity | ||
==Machine learning approaches to evolution (Led by Oliver & Brian)== | ==Machine learning approaches to evolution (Led by Oliver & Brian)== | ||
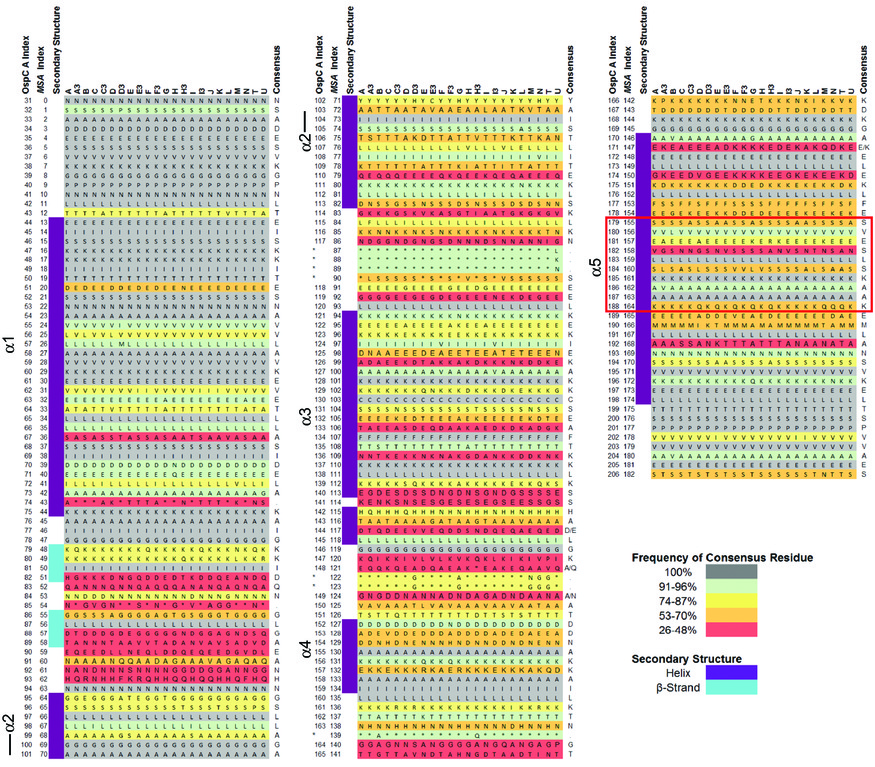
[[File:S2-lo-res0.jpg|thumbnail]] | [[File:S2-lo-res0.jpg|thumbnail|Image converted from S2 from Baum et al (2013)]] | ||
# Goal 1. Implement Hopfield network for optimization of protein structure | # Goal 1. Implement Hopfield network for optimization of protein structure | ||
# Goal 2. Neural-net models of OspC. Structural alignment (S2 from Baum et al 2013): | # Goal 2. Neural-net models of OspC. Structural alignment (S2 from Baum et al 2013): | ||
Revision as of 17:36, 16 June 2018
Rules of Conduct
- No eating, drinking, or loud talking in the lab. Socialize in the lobby only.
- Be respectful to each other, regardless of level of study
- Be on time & responsible. Communicate in advance with the PI if late or absent
Participants
- Dr Oliver Attie, Research Associate
- Brian Sulkow, Research Associate
- Saymon Akther, CUNY Graduate Center, EEB Program
- Lily Li, CUNY Graduate Center, EEB Program
- Mei Wu, Bioinformatics Research Assistant
- Yinheng Li, Informatics Research Assistant
- Christopher Panlasigui, Hunter Biology
- Dr Lia Di, Senior Scientist
- Dr Weigang Qiu, Principal Investigator
- Summer Interns: Mahmad, Pavan, Roman, Benjamin, Andrew, Michelle, Hannah
Journal Club
- a Unix & Perl tutorial
- A short introduction to molecular phylogenetics: http://www.ncbi.nlm.nih.gov/pubmed/12801728
- A review on Borrelia genomics: https://www.ncbi.nlm.nih.gov/pubmed/24704760
- ospC epitope mapping: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0067445
Projects
Borrelia genome evolution (Led by Saymon)
- Goal 1. Estimate time of cross-Atlantic dispersal using core-genome sequences
- Goal 2. Investigate codon biases with respect to levels of gene expression. Data file:
Identification of host species from ticks (Led by Lily [after first-level])
- Goal 1. Protocol optimization for PCR amplification of host DNA from ticks
- Goal 2. Protocol development: library construction for MiSeq
- Goal 3. Development of bioinformatics protocols and sequence database
Pseudomonas Genome-wide Association Studies (GWAS) (Led by Mai & Yinheng, in collaboration with Dr Xavier of MSKCC)
- Goal 1. Association of genes/SNPs with biofilm formation and c-di-GMP levels: Manuscript preparation
- Goal 2. Association of genome diversity with metabolic diversity
Machine learning approaches to evolution (Led by Oliver & Brian)
- Goal 1. Implement Hopfield network for optimization of protein structure
- Goal 2. Neural-net models of OspC. Structural alignment (S2 from Baum et al 2013):
- Goal 3. K-mer-based pipeline for genome classification
Weekly Schedule
- Summer kickoff (June 1, 2018, Friday): Introduction & schedules
- Week 1 (June 4-8):
- Monday: the Unix & Perl Tutorial, Part 1
- Tuesday: Unix Part2. Explore the "iris" data set using R, by following the the Monte Carlo Club Week 1 (1 & 2) and Week 2. Read McKay (2003), Chapters 38 & 39
- Thursday: 1st field day (Caumsett State Park); Participants: John, Mahamad, Pavan, Andrew, Dr Sun, Weigang, with 3 members of Moses team from Suffolk County Vector Control. Got ~110 deer tick nymphs
- Friday: meeting with MSKCC group at 11am; BBQ afterwards
- Week 2 (June 11-15):
- Monday: Lab meeting, projects assigned
- Tuesday: neural net tutorial (by Brian)
- Thursday: 2nd field day (Fire Island National Seashore). Participants: John, Brian, Mei, Mahamad, Pavan, Benjamin, and Weigang. Got ~100 lone-star ticks and 4 deer tick nymphs
- Week 3 (June 18-22):
- Monday: Lab meeting, 1st project reports
- Codon Bias: Theory, Coding, and Data (Andrew, Pavan, Saymon)
- OspC epitope identification: Serum correlation, sequence correlation, immunity-sequence correction (Mahamad, Roman, Brian)
- Pseudomonas metabolomics: parsing intensity file; theory & parsing SMBL file (Chris & Benjamin)
- Monday: Lab meeting, 1st project reports
- June 29 (Friday)
- July 5 (Thursday)
- July 12 (Thursday, by skype)
- July 19 (Thursday, by skype)
- July 26 (Thursday, by skype)
Lab notes for Summer HS Interns
- NCI Cloud: Seven Bridges Cloud Platform. Create an user account
- Read documentation & tutorials: Documentation
Notes & Scripts
- (Weigang) A sample R script to parse Table S2 from Baum et al 2013, sera-antigen reactivity measurements
# preliminaries: save as TSV; substitute "\r" if necessary;
# substitute "N/A" to "NA"; remove extra columns
setwd("Downloads/")
x <- read.table("table-s2.txt4", sep="\t", header=T)
View(x)
colnames(x)
which(x[,8]=="A")
x[which(x[,8]=="A"),12]
x[which(x[,8]=="A3"),12]
cor.test(x[which(x[,8]=="A3"),12], x[which(x[,8]=="A"),12], method = "pearson")
x.cor$estimate
levels(x[,8]) # obtain ospC allele types; to be looped through in pairwise
for (i in 1:?) { for (j in ?:?) {cor.test(....)}}