Biol375 2018: Difference between revisions
imported>Weigang |
imported>Weigang |
||
| Line 107: | Line 107: | ||
| | | | ||
# Download or Copy/Paste [http://media.hhmi.org/biointeractive/activities/lizard/Anolis-DNA-sequences.txt the lizard DNA sequences] to your own computer and save the file as "lizard.txt" | # Download or Copy/Paste [http://media.hhmi.org/biointeractive/activities/lizard/Anolis-DNA-sequences.txt the lizard DNA sequences] to your own computer and save the file as "lizard.txt" | ||
# Align the DNA sequences [http://www.phylogeny.fr/one_task.cgi?task_type=muscle using this website] and save the aligned DNA file ("Output->Alignment in Fasta format") as "lizard-aligned.txt" | # Align the DNA sequences [http://www.phylogeny.fr/one_task.cgi?task_type=muscle using this website] and save the aligned DNA file ("Output->Alignment in Fasta format") as "lizard-aligned.txt". Use "one-click" option to make a tree. | ||
# Based on [http://media.hhmi.org/biointeractive/activities/lizard/Lizard-Cards-Color.pdf the lizard card], construct a character-state matrix for all lizard species. For each species, list its character state for each of the following two characters (as columns): (1) Geographic origin, and (2) Habitat. | # Based on [http://media.hhmi.org/biointeractive/activities/lizard/Lizard-Cards-Color.pdf the lizard card], construct a character-state matrix for all lizard species. For each species, list its character state for each of the following two characters (as columns): (1) Geographic origin, and (2) Habitat. | ||
# Determine which hypothesis ("Multiple origin" or "Single origin" of ecomorphs) is more supported by the mtDNA tree. Explain. | |||
|} | |} | ||
* 10/1 (M). Parsimony reconstruction (Chapter 5). | * 10/1 (M). Parsimony reconstruction (Chapter 5). | ||
Revision as of 21:12, 27 September 2018
Course Description
Molecular evolution is the study of the change of DNA and protein sequences through time. Theories and techniques of molecular evolution are widely used in species classification, biodiversity studies, comparative genomics, and molecular epidemiology. Contents of the course include:
- Population genetics, which is a theoretical framework for understanding mechanisms of sequence evolution through mutation, recombination, gene duplication, genetic drift, and natural selection.
- Molecular systematics, which introduces statistical models of sequence evolution and methods for reconstructing species phylogeny.
- Bioinformatics, which provides hands-on training on data acquisition and the use of software tools for phylogenetic analyses.
This 3-credit course is designed for upper-level biology-major undergraduates. Hunter pre-requisites are BIOL203, and MATH150 or STAT113.
Please note that starting from fall 2015, completing this course no longer counts towards research credits for biology majors.
Textbooks
- (Required) Graur, 2016, Molecular and Genome Evolution, First Edition, Sinauer Associates, Inc. ISBN: 978-1-60535-469-9. Publisher's Website (Student discount: a 15% discount and receive free UPS standard shipping)
http://www.sinauer.com/molecular-and-genome-evolution.html)
- (Recommended) Baum & Smith, 2013. Tree Thinking: an Introduction to Phylogenetic Biology, Roberts & Company Publishers, Inc.
Learning Goals
- Be able to describe evolutionary relationships using phylogenetic trees
- Be able to use web-based as well as stand-alone software to infer phylogenetic trees
- Understand mechanisms of DNA sequence evolution
- Understand algorithms for building phylogenetic trees
Links for phylogenetic tools
- NCBI sequence databases
- R Tools
- R source: download & install from a mirror site
- R Studio: download & install
- APE package
- A Molecular Phylogeny Web Server
- EvolView: an online tree viewer
Exams & Grading
- Bonus for full attendance & active participation in classroom discussions.
- Assignments. All assignments should be handed in as hard copies only. Email submission will not be accepted. Late submissions will receive 10% deduction (of the total grade) per day.
- Three Mid-term Exams (30 pts each)
- Comprehensive Final Exam (50 pts)
Academic Honesty
While students may work in groups and help each other for assignments, duplicated answers in assignments will be flagged and investigated as possible acts of academic dishonesty. To avoid being investigated as such, do NOT copy anyone else's work, or let others copy your work. At the least, rephrase using your own words. Note that the same rule applies regarding the use of textbook and online resources: copied sentences are not acceptable and will be considered plagiarism.
Hunter College regards acts of academic dishonesty (e.g., plagiarism, cheating on examinations, obtaining unfair advantage, and falsification of records and official documents) as serious offenses against the values of intellectual honesty. The College is committed to enforcing the CUNY Policy on Academic Integrity and will pursue cases of academic dishonesty according to the Hunter College Academic Integrity Procedures.
Course Schedule
Part 1. Tree Thinking
- 8/27 (M). Overview & Introduction. Textbook Chapter: "Introduction" (pages 1-3)
| Assignment 1 (10 pts; Due next class 8/30) |
|---|
|
- 8/30 (TH). Introduction (Continued).
- Go over pre-test questions
- Tutorial: R & R-Studio (Bring your own computer).
| Assignment 2 (5 pts; Due: next session) |
|---|
R exercises
|
- 9/5 (Wed; Monday Schedule). Intro to trees.
- In-class exercise 1. (5 pts; Due next session)
- Introductory slide:
- 9/6 (TH). Intro to trees.
- In-class exercise 2. (5 pts; Due next session)
- Textbook Chapter 5: "Molecular Phylogenetics" (pages 170-175; 201-202)
- 9/13 (TH). Species Tree & Lineage Sorting.
- Textbook Chapter 5: "Molecular Phylogenetics" (pages 177-180).
- 9/17 (M). Consensus Tree & Review.
- Chapter 5. pages 199-200 (Figure 5.31)
- In-class exercise 3. (5 pts, due next session)
- Lecture Slides:
- 9/20 (Th). 4:10 - 5:10pm Midterm Exam I Bring pencils, erasers, and a calculator
Part 2. analysis of Trait Evolution
- 9/24 (M). Traits & trait matrix
- Textbook Chapter 5, pages 180-183
| Assignment #3 (5 pts; Due next session) |
|---|
Watch Origin of Species: Lizards in an Evolutionary Tree. Provide short answer (1-3 sentences) to each of the following three questions.
|
- 9/27 (TH). Homoplasy & consistency
| Assignment #4 (5 pts; Due next session) |
|---|
|
- 10/1 (M). Parsimony reconstruction (Chapter 5).
- Textbook Chapter 5, pages 188-191
- In-Class Exercise 4
- 10/4 (TH). Genome & gene structure (Chapter 3)
- In-Class Exercise 5. Pretest Part 2 (molecular phylogenetics in forensics)
- 10/11 (TH). In-class exercises
- 10/15 (M). Review & Practices. Lecture Slides:
- 10/18 (TH). Midterm Exam 2
Part 3. Tree Algorithms
- 10/22 (M). BLAST & Alignments (Chapter 3. pages 93-100)
| Assignment #6 (10 pts; Due 10/30, Monday) |
|---|
Based on the NCBI Gene Page for cytochrome C (CYCS), answer the following questions:
|
- 10/25 (TH). Genetic distances & Sequence-evolutionary models (Chapter 3, pages 79-88)
- 10/29 (M). Maximum parsimony (Chapter 5, pages 191-194).
- 11/1 (TH). Distance methods (Chapter 5, pages 184-187). In class exercise #6; In-class computer exercise:
| Assignment #7 (10 pts). Due 11/6 (Monday) |
|---|
|
- 11/5 (M). Likelihood & Bayesian methods (Chapter 5, pages 194-198). Lecture slides posted:
In-class exercise:
| Assignment #8 (10 pts). Due 11/13 (Monday) |
|---|
|
- 11/8 (TH). Tree-testing & Review (Chapter 5, pages 207-209). Lecture slides posted in the last session
- 11/12 (M). Midterm Exam 3
Part 4. Population Genetics (Chapter 2)
- 11/15 (TH). Mechanism of molecular evolution: Overview (pages 35-38); SNP statistics
- 11/19 (M).
| Assignment #9 (10 pts; Due 11/30, Thursday) |
|---|
|
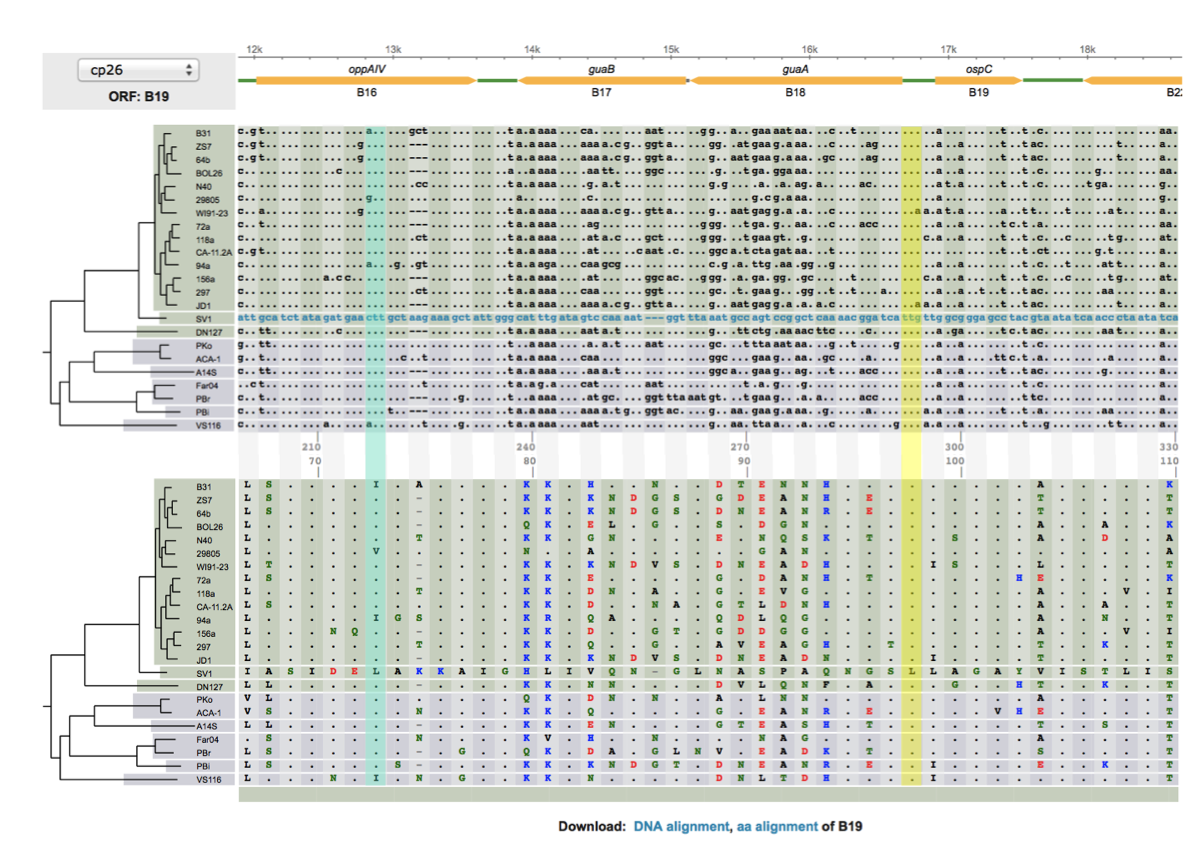
The left figure shows a codon alignment of 38 strains of a bacterium, with an outgroup sequence (which starts with a string of SNPs: "....g...c..ca..", etc), answer the following questions (with the outgroup sequence excluded.) Do not print the figure directly. Hand-copy the sequences to a graph sheet, include only sequences at the two variable codon positions:
|
- 11/26 (M).
- 11/29 (TH). Neutral Theory & Molecular Clock (pages 58-59; 72-74)
- 12/3 (M). Genetic Drift (pages 47-49). Computer exercises of Assignment #10 (below) will be done in class.
| Assignment # (10 pts; Due 12/11, Monday) |
|---|
Statistical experiments to explore gene-frequency change due to genetic drift:
|
- 12/6 (TH) Rates of nucleotide substitutions (pages 111-125). Part 4 slides:
- 12/11 (M). (Last Lecture) Review & Course evaluations. Review slides: . Submit your Teacher's Evaluation, using either:
- Personal computer at www.hunter.cuny.edu/te; or,
- Smartphone at www.hunter.cuny.edu/mobilete
- 12/17 (Monday, 4-6pm) Comprehensive Final Exam