A Primer on the Cluster System at Hunter
What is a Cluster System?
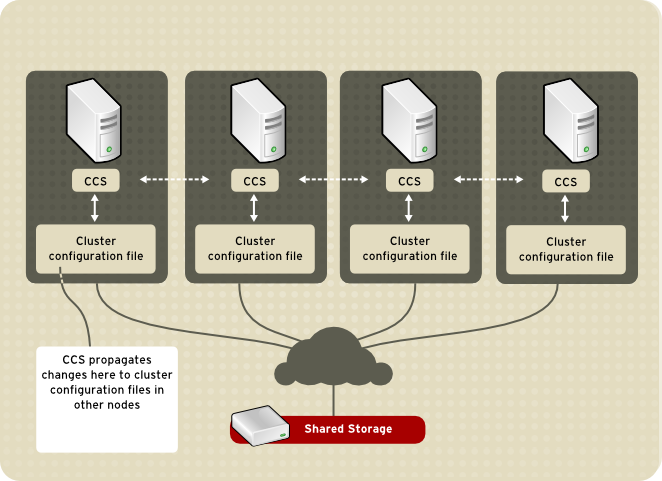
A Cluster System is a set of connected computers that work together in order to perform tasks, unlike many of the portals at hunter that require you to first login to a head node, and then to a working node.
- For example to enter the Qiu lab servers you must first login to Darwin.hunter.cuny.edu and then to a compute node such as Wallace.hunter.cuny.edu
On a cluster system all of the nodes connected can be viewed as a single system. Under this system all you would need to do is login to the head node, and from there you can run your programs. However, unlike the non-cluster computer servers at hunter, in order to fully utilize the cluster you must submit your job to a job scheduler that controls which node to run the job on.
Basics of Using a Cluster System
- What programs are currently installed on the cluster System?
The normal way of running programs on a linux machine would be to type the name of the program into the terminal, add any options to the command, and press enter. For example"
$>blastp -query test.fas -db nr -remote -outfmt 6However this will not work in a cluster. In order to run programs on a cluster, the cluster must first have those programs installed (or you may have to compile the program and run it locally) and then you must load the program on to your terminal. To check the available programs on the Hunter College cluster use this command:
module availThis command shows what modules are currently available to use. To save the list of available programs use this command:
module avail 2> ~/available_apps.txtThis command stores the present list programs installed on the cluster to a file called "available_apps.txt" to your home directory. This file will only show the present programs installed on the cluster, you will need to update this file in the future to show new applications!
- How do we load a program on the cluster?
Now that we know what are the available apps on the cluster, we need to load the application into our terminal. To do so we use the command:
module load <Program Name>For example, to load blast on the cluster we need to run the command:
module load R/3.2.2Notice how we have a version number when we run the module load command? This means that on the cluster we can use different versions of a program! This is useful for those situations when an updated version of a program breaks compatibility with some code you've written or some modules you've downloaded, or if you like one version of a program more than the other.
In order to use a different version of a program we first need to unload our program then load up the correct version of the program:
module unload R/3.2.2
module load R/3.1.0- How do we run a program on the cluster?
Now lets go back to our previous blast example. Lets first load up the module:
module load blast/2.2.31Notice now, when you tab "bl" on the command line all programs associated with blast comes up, such as blastn / blastp, etc. Now you may be tempted to run blast, or any program, the normal way, that is:
blastp -query test.fas -db nr -remote -outfmt 6Don't do this!!! When you run the command like this, you are running it on the head node!! You aren't using the power of the cluster to do the work, that and you may be clogging up the little computational power (relative to the compute nodes) the head node has and making everyone else experience on the cluster laggy and unstable! To run a job on the cluster you must run it through a scheduler, in our case the cluster uses SLURM, but the same principals apply to other cluster formats such as LSF or BSUB.
The correct way of submitting a job to the cluster from the terminal is to use this command:
srun <command>>This will submit the command to a compute node, and run the job interactively, that is, all output will redirect to the terminal you are using, for example:
module load cdhit/4.64
srun cd-hit -hWill output the help directly to your terminal. While this is fine for quick and dirty programs like cdhit, when you run blast you'll get a blank output until the blast is complete. Try it for your self:
blastp -query test.fas -db nr -remote -outfmt 6If you close the terminal, or if you lose your connection to the internet during this time, all progress will be lost. So how do we submit jobs to the cluster and just let things run without worrying about our job dying? Also can we allocate specific resources to our programs, like the number of cpu's it needs, or the amount RAM? To do this we need to write some scripts, so open up a text editor and head to the next section, yo.
Cluster Scripting
In order to submit a job to the cluster and have it run without keeping a terminal open we need to write a script. You can use this template script as a basis for any future cluster scripts you may need to write:
#!/bin/bash
#
#SBATCH --job-name=<job name>
#SBATCH --output=<out name>
#
#SBATCH --ntasks=<number of tasks>
#SBATCH --time=<time needed to run task>
#SBATCH --mem-per-cpu=<memory per cpu needed>
#SBATCH --mem-per-cpu=<memory per cpu needed>
#SBATCH --cpus-per-task=<number of cpus>
<Bash scripting goes here >This is basically what you need to submit a job to the cluster system we have at hunter. Now lets go through it, shall we?
#SBATCH --job-name=<job_name>This line of code allows you to specify the name of your job, for example if I want my job name to be "pandas_are_awesome" then I would use the following line:
#SBATCH --job-name=pandas_are_awesomeThe next line of code let you specify the file you want to dump all output (both stdout and stderr) to the terminal into. Remember we are not in interactive mode here, so we wont see any terminal output. So if we want to dump the output to pandas_are_not_awesome.log we would use the following line:
#SBATCH --output=pandas_are_not_awesome.logHopefully the rest of the #SBATCH lines are self explanatory. After we put in what is essentially metadata about our job, we can then write a plain bash script to run our job. So if I wanted to run blast I would use the following code:
#!/bin/bash
#
#SBATCH --job-name=blasttest
#SBATCH --output=blasttest.log
#
#SBATCH --ntasks=1
#SBATCH --time=10:00 #10 minutes
#SBATCH --mem-per-cpu=100 #megabytes
#SBATCH --cpus-per-task=4
module load blast/2.2.31 ##load the module
blastp -query test.fas -db nr -remote -outfmt 6 -out blasttest.out ##run the programNotice two things:
- I needed to load the blast module inorder for my code to run, you need to do this if your program needs to be loaded in order for it to run.
- You did not have to write "srun" before the command.
Since after the #SBATCH lines, the script becomes a bash scrips, we can also use loops. For example say I had a bunch of sequences in a folder that I wanted to blast; I would use this code:
#!/bin/bash
#
#SBATCH --job-name=blasttest
#SBATCH --output=blasttest.log
#
#SBATCH --ntasks=1
#SBATCH --time=10:00 #10 minutes
#SBATCH --mem-per-cpu=100 #megabytes
$SBATCH --cpus-per-task=4
module load blast/2.2.31 ##load the module
for file in ./*.fasta ##go through every file that ends in .fasta
do
name=$(basename $file ".fasta" ) ## get the file name
blastp -query $file -db nr -remote -outfmt 6 -out $name.out ##run blast on the current file
doneCool beans? Now, after we finish writing and saving the script, how do we run it exactly? To do this we use the command on the terminal:
$> sbatch <script.sh>So if the above script was called "test.sh" to run it I would use the command:
$> sbatch test.shAnd it should run without any problems and is now safe to close the connection to the cluster. If we wanted to check on our job there are two possible ways to do it: the not so cool way of looking at our output file we specified in the script. (In this case blasttest.log ) or the cooler ways seen here:
Monitoring Cluster Jobs
To check if your cluster job is running we use the command:
$> squeueOur output would be something like this:
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
164 defq bash root R 17:45:29 1 compute003
165 defq bash root R 17:45:11 1 compute004
168 defq bash averdes R 17:42:19 1 compute006
163 himem bash jgorson R 1-17:51:33 1 compute018
170 himem bash rrahman blastp 1-17:51:33 1 compute018Notice, we can not only see our own jobs, but also the jobs of the other people on the cluster, so if you are doing something that is incredibly computationally intensive and slowing everyone else down, we will see it and we will bring pitchforks. We can also use the command:
$> sstatTo know whats going on with our job, like how much ram or cpu threads it's using, etc.
If we want to kill our job, for whatever reason, we would use the command:
$> scancel <jobid>To get our job id, we use squeue, its the first column.
Cluster File System and General Information
To login to the cluster we ssh into 146.95.252.94. This can only be done within Hunter, not outside of the firewall.
The cluster file system is separated between 3 areas for storing files and data:
- Scratch: Scratch space on a file system refers to a shared allocation for all users in the cluster system. Scratch space is reserved for temporary files. The amount of space allocated here is virtually unlimited however files in the scratch directory are purged (deleted) every month.
- The directory for the scratch space in Hunters Cluster System is:
/scratch/- Personal: You have access to about 10GB of space in your home directory. However since it is your home directory on the cluster on one else can edit or read your files. Files here are not purged
- The directory for your personal space in Hunters Cluster System is:
~/
or
/home/<username>- Projects: The project directory is the directory where you should store files related to the work you are doing. Here everyone in your group can view and edit files. Additionally your group has allocations to about 1TB of storage space, collectively. Finally Files here are not purged
- The directory for your projects space in Hunters Cluster System is:
/lustre/projects/<groupname>
in our case this is
/lustre/projects/qiulabConclusion
Hopefully after following this tutorial you have risen from cluster Padawan, to cluster Jedi. But complete, your training is not. There's always something new you can learn about using the cluster system from google or stack overflow. If you find something new or interesting that you think might be useful for other people to use email me at rayees.rahman40@myhunter.cuny.edu and I can add it to this write up.
This write up is far from complete (we don't even have a name for our cluster yet, come on!) so new things will be added all the time, so watch this page!
Rayees
Conda Usage
# to create an environment
module load anaconda
conda create --name gbs python=3.10 bwa samtools
conda info --envs
# to run
module load anaconda
source activate gbs
Pre-written cluster scripts for various software
Here are examples of scripts written for the cluster that use various software, feel free to use these script to learn how to run particular pieces of bioinformatics software or to create more expansive genomics pipelines. Also please feel free to contribute to this page! Send me or Dr. Qiu an email to update this section with new example scripts for running software.
BWA & Samtools
Use array option to loop through directories and files
#!/bin/bash
#
#SBATCH --job-name=gbs
#SBATCH --output=gbs.log
#SBATCH --array=1-2
#SBATCH -o output-%A_%a-%J.o
#SBATCH -n 1
#
# Reference for array operations: https://portal.supercomputing.wales/index.php/index/slurm/interactive-use-job-arrays/job-arrays/
# formal doc: https://slurm.schedmd.com/job_array.html
# %J: job identifier
# %A: parent job
# %a: array iteration index
work_dir=/home/wqiu/project-home/GBS-April-2023
module load anaconda
source activate gbs
echo SLURM_JOB_ID $SLURM_JOB_ID
echo SLURM_ARRAY_JOB_ID $SLURM_ARRAY_JOB_ID
echo SLURM_ARRAY_TASK_ID $SLURM_ARRAY_TASK_ID
DIRS=(GBS100 GBS101)
for i in ${DIRS[@]}
do
echo "Processing folder $i"
dir=$work_dir/$i
bwa mem $work_dir/ref.fas $dir/$i.R1.fq.gz $dir/$i.R2.fq.gz > $dir/$i.sam 2> /dev/null
echo "sam file generated: $i.sam"
samtools view -bT $work_dir/ref.fas $dir/${i}.sam | samtools sort -o $dir/${i}.bam 2> /dev/null
echo "bam file generated: $i.bam"
doneInterproscan
Change the wall clock limit to whatever you feel is appropriate, interproscan runs, as far as I can tell, using only a single core, so it will be pretty slow. Let me know if we can improve the speed of it. -Rayes
#!/bin/bash
#
#SBATCH --job-name=interproscan
#SBATCH --output=interpro.log
#
#SBATCH --ntasks=1
#SBATCH --time=4320:00
#SBATCH --mem-per-cpu=3000
#SBATCH --cpus-per-task=4
module load interproscan/5.14.53.0
interproscan.sh -i cdhit-40 -o cdhit-40-out.txt -t p -goterms -pa -f tsvVelvet
Change the wall clock limit to whatever you feel is appropriate, interproscan runs, as far as I can tell, using only a single core, so it will be pretty slow. Let me know if we can improve the speed of it. -Rayes
#!/bin/bash
#
#SBATCH --job-name=interproscan
#SBATCH --output=interpro.log
#
#SBATCH --ntasks=1
#SBATCH --time=4320:00
#SBATCH --mem-per-cpu=3000
#SBATCH --cpus-per-task=24
module load velvet/1.2.10
velveth ./test2 21 -fastq -long ./N18_S15_L001_R1_001_mat.fq
velvetg ./test2 -cov_cutoff 4 -min_contig_lgth 100Setting up a conda/mamba environment on the HPC
Create a space for your installation
Use the project space since the quota enforced on your home directory is quite small.
mkdir -p /lustre/projects/qiulab/$USER
Setting up mambaforge
Follow the instructions on the [miniforge git repository](https://github.com/conda-forge/miniforge#mambaforge) (partially reproduced below):
curl -L -O "https://github.com/conda-forge/miniforge/releases/latest/download/Mambaforge-$(uname)-$(uname -m).sh"
bash Mambaforge-$(uname)-$(uname -m).sh
During the setup wizard, you will be asked to enter the directory in which to install Mambaforge. Enter the directory you created above and add "/mambaforge" to the end. For example, if your username is "user", you would enter:
/lustre/projects/qiulab/user/mambaforge
Replace "user" above with your own username to use the directory created earlier.
At the end of the installation, you will be asked whether or not you would like to run `conda init`. Answer "yes". When complete, close and reopen your shell (or disconnect/reconnect to the HPC node).
Configuring bioconda
Bioconda(https://bioconda.github.io/) is a conda repository for software used in biology. You'll need to add this repository for most of the tools you will be using.
Check the website for the latest instructions, but the commands to add the repository (as of Jun, 2023) are below:
conda config --add channels defaults
conda config --add channels bioconda
conda config --add channels conda-forge
conda config --set channel_priority strict
Creating a new environment
You can create environments using mamba create (or conda create). An environment can be used to install all of the software needed to complete some task or workflow, without interfering with the system installation. You will still be able to access software installed to your operating system's default PATH, but software in an environment (usually) takes precedent, and is isolated to that environment. In other words, if you install a copy of clustalw in a conda environment, and you activate that environment, it will (should) mask whatever version of clustalw you may have installed in your system. Also, you can have more than one version of clustalw installed simultaneously, each one being installed in its own environment.
From scratch
You can create a new enviroment using mamba create and supplying a name for the environment. Optionally (but strongly recommended), you can end the command with a list of packages. e.g.,
mamba create aligners -n blast clustalo muscle bwa minimap2 mafft
This will create a new environment called "aligners" with blast, clustalo, muscle, bwa, minimap2, and mafft available for use once activated. To activate an environment, do:
mamba activate aligners
Post-environment creation, you can install packages using the install subcommand:
mamba install -n aligners diamond
You can also install specific versions of a software package:
mamba install -n aligners blast=2.10
The above will get conda/mamba to try to downgrade the package, if it is already installed. This may or may not work if the system cannot resolve any dependency issues that may occur with dependent packages.
From an environment.yml file
Conda users can share their environments in a fairly convenient fashion by dumping the list of packages:
conda env export -n aligners > aligners_environment.yml
This will export the list of packages installed in the `aligners`, along with their exact version, into a YAML file called aligners_environment.yml. You can share this file with others and conda/mamba will attempt to recreate the environment on their machine.
To import a YAML file you recieved, run:
mamba env create -f environment.yml
This will create a new environment with whatever name was specified in the environment.yml file. To choose your own, add -n envname.
Using conda/mamba
See the "From scratch" section above for a few examples.
Installing Qiu-lab software
mamba create -n qiulab perl-bio-bpwrapper