EEB BootCamp 2020
Jump to navigation
Jump to search
| Lyme Disease (Borreliella) | CoV Genome Tracker | Coronavirus evolutuon |
|---|---|---|
Case studies from Qiu Lab
CoV genome data set
- N=565 SARS-CoV-2 genomes collected during January & February 2020. Data source & acknowledgement GIDAID (Warning: You need to acknowledge GISAID if you reuse the data in any publication)
- Download file: data file
- Create a directory, unzip, & un-tar
mkdir QiuAkther
mv cov-camp.tar.gz QiuAkther/
cd QiuAkther
tar -tzf cov-camp.tar.gz # view files
tar -xzf cov-camp.tar.gz # un-zip & un-tar
- View files
file TCS.jar
ls -lrt # long list, in reverse timeline
less Jan-Feb.mafft # an alignment of 565 CoV2 genomes in FASTA format; "q" to quit
less cov-565strains-617snvs.phy # non-gapped SNV alignment in PHYLIP format
wc hap.txt # geographic origins
head hap.txt
wc group.txt # color assignment
cat group.txt
less cov-565strains.gml # graph file (output)
Bioinformatics Tools & Learning Goals
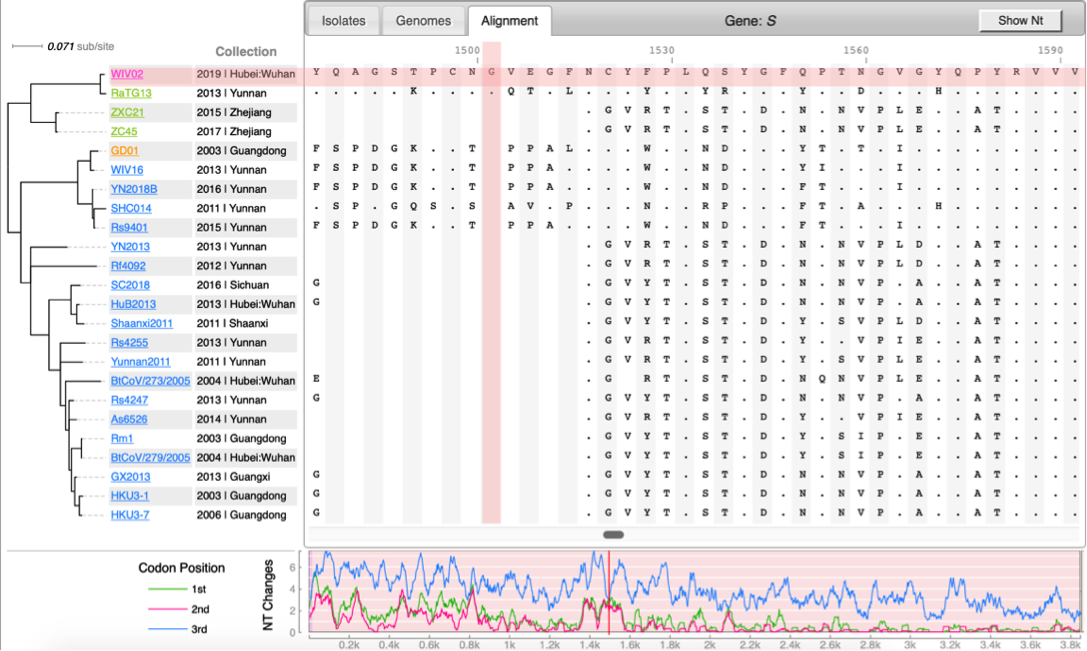
- BpWrapper: commandline tools for sequence, alignment, and tree manipulations (based on BioPerl).
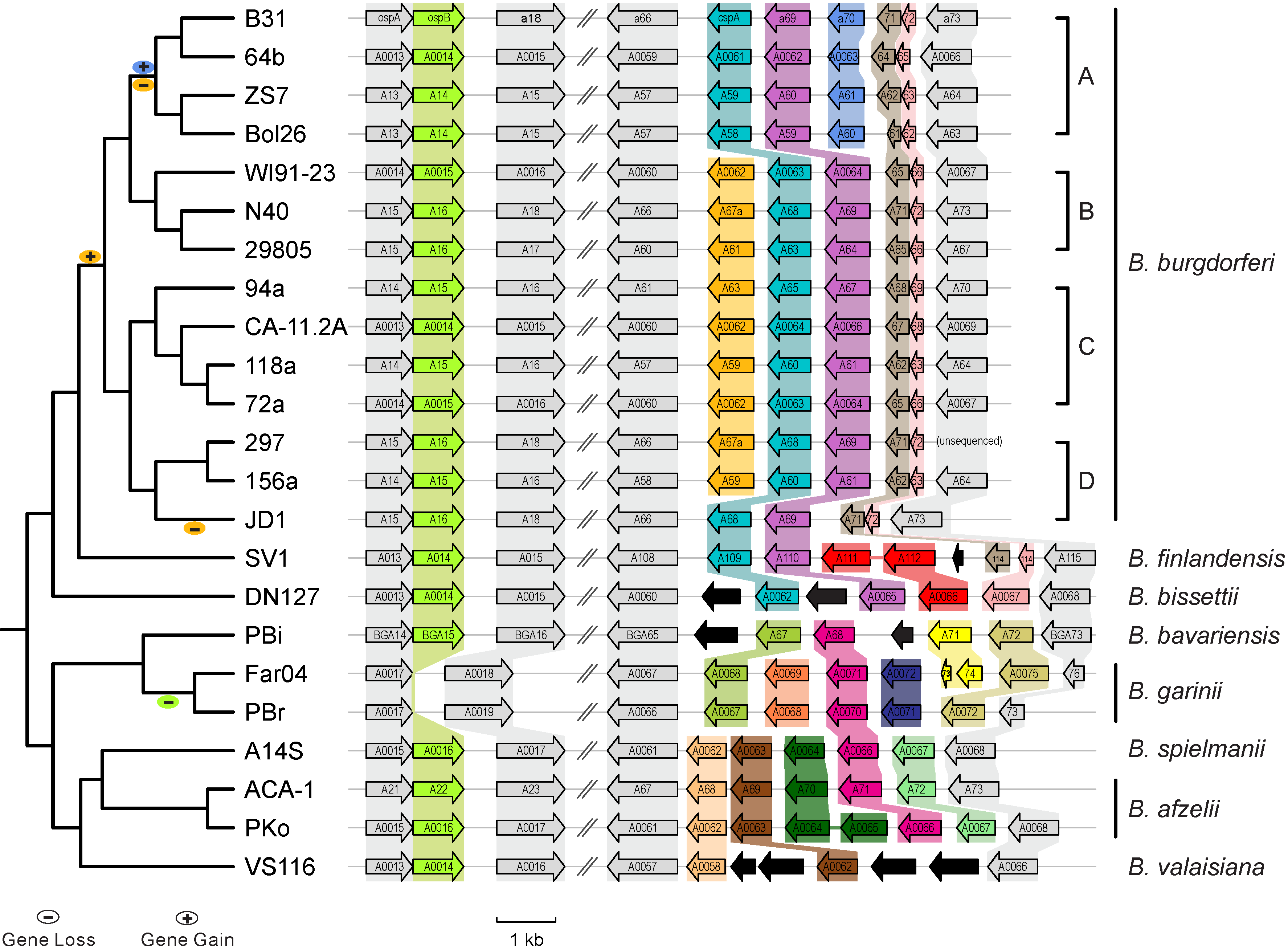
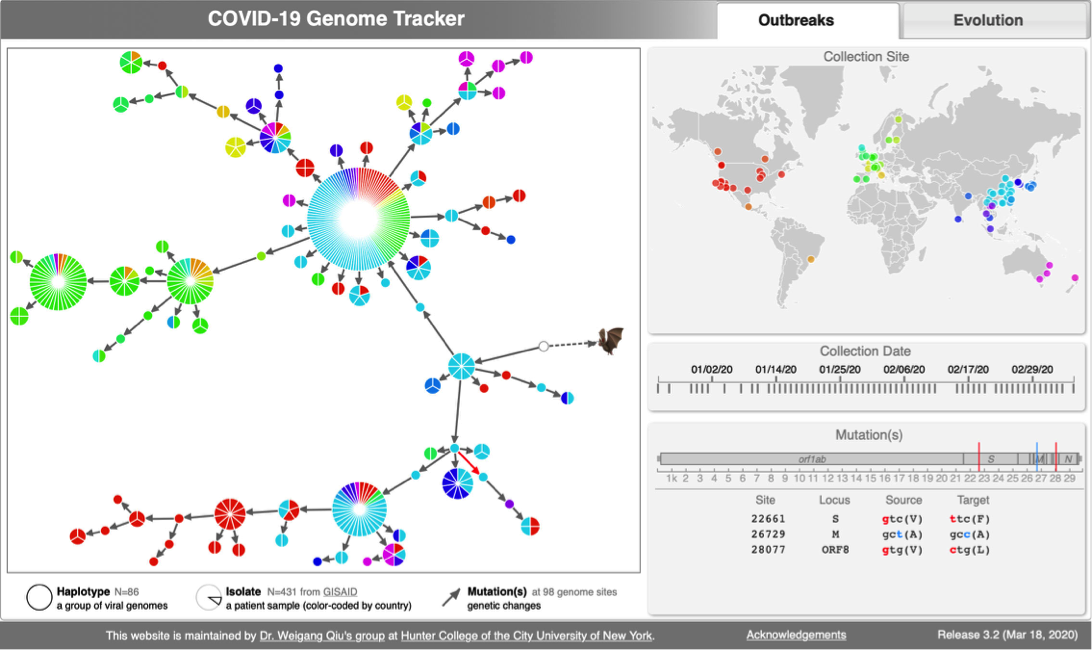
- Haplotype network with TCS PubMed link
- Web-interactive visualization with D3js
Tutorial
- 2-2:30: Introduction on pathogen phylogenomics
- 2:30-2:45: Demo: sequence manipulation with BpWrapper
<syntaxhighlight lang='bash'> bioseq --man bioseq -n Jan-Feb.mafft bioaln --man bioaln -n -i'fasta' Jan-Feb.mafft bioaln -l -i'fasta' Jan-Feb.mafft bioaln -n -i'phylip' cov-565strains-617snvs.phy bioaln -l -i'phylip' cov-565strains-617snvs.phy FastTree -nt cov-565strains-617snvs.phy > cov.dnd biotree --man biotree -n cov.dnd biotree -l cov.dnd <syntaxhighlight>
- 2:45-3:00: build haplotype network with TCS
<syntaxhighlight lang='bash'> java -jar -Xmx1g TCS.jar <syntaxhighlight>
- 3:00-3:15: interactive visualization with BuTCS
- 3:15-3:30: Q & A