BorreliaBase Help
What is BorreliaBase
How to Cite BorreliaBase
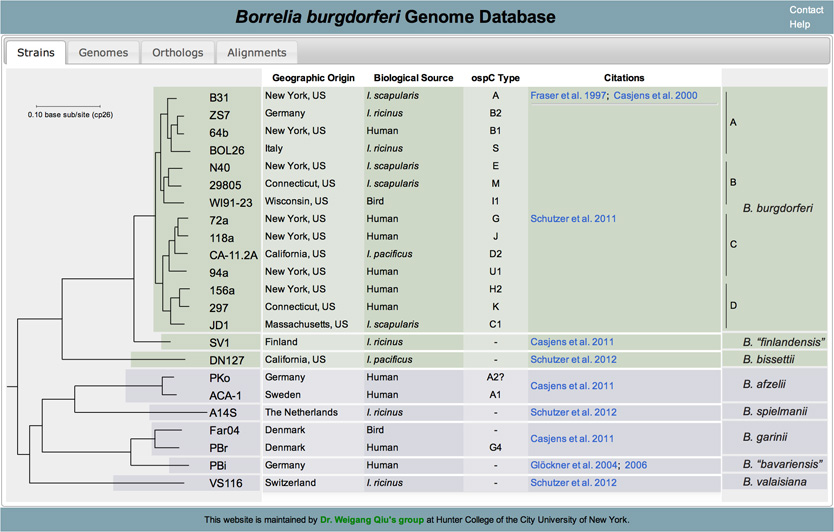
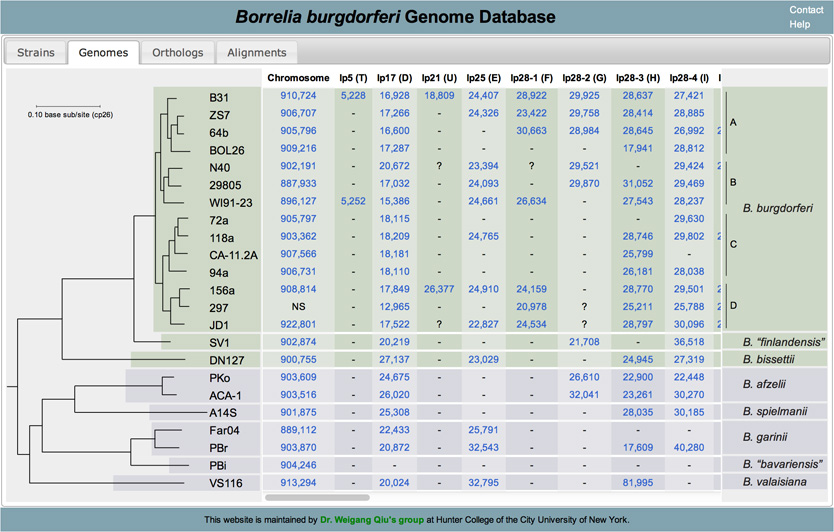
Current list of genomes
Current list of orthologs
Tutorial 1. How to retrieve orthologous ORF sequences?
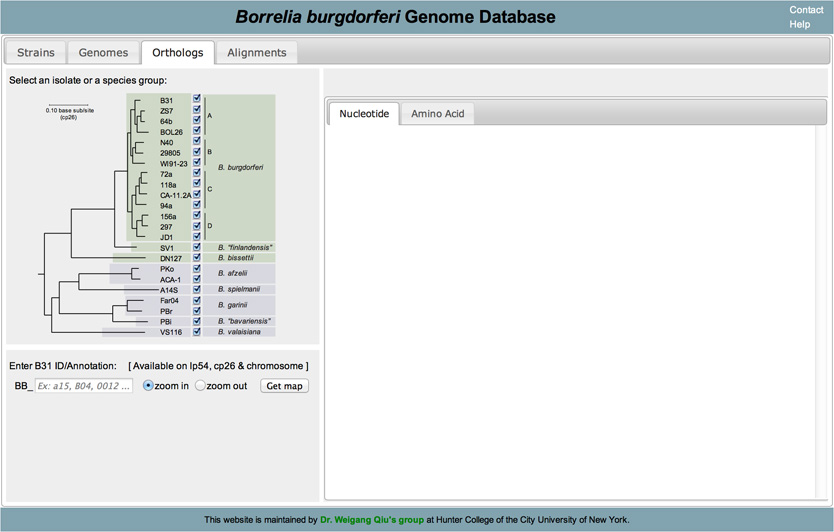
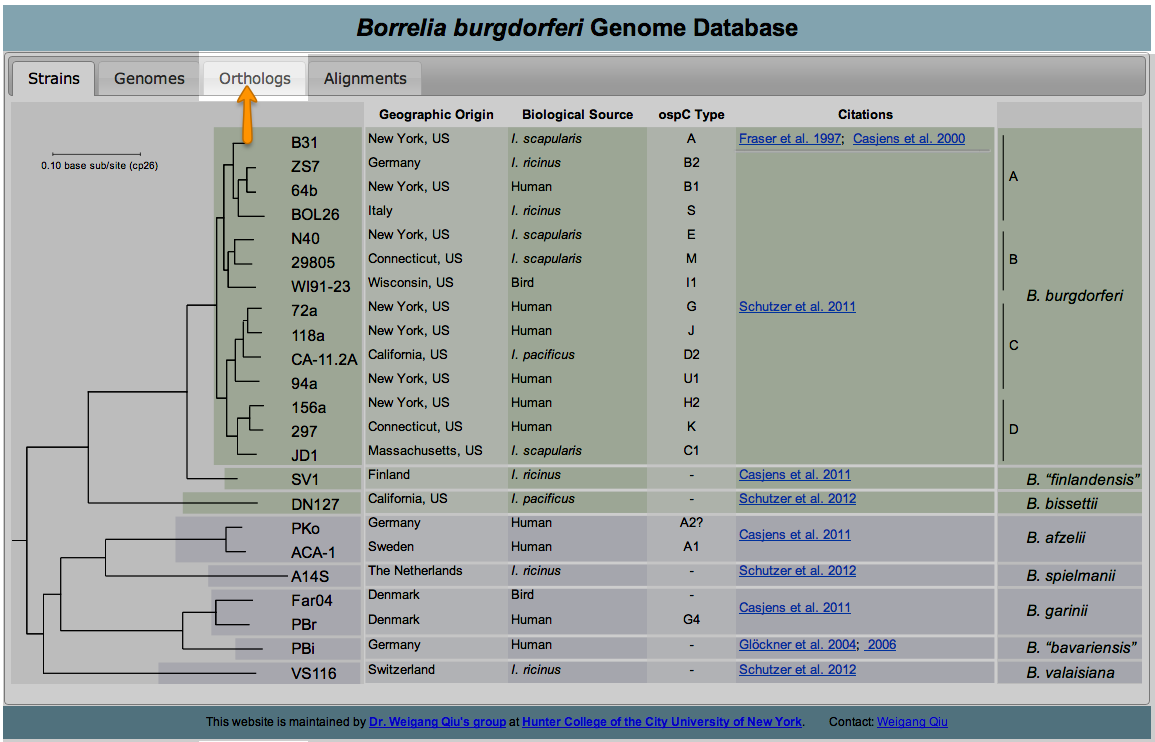
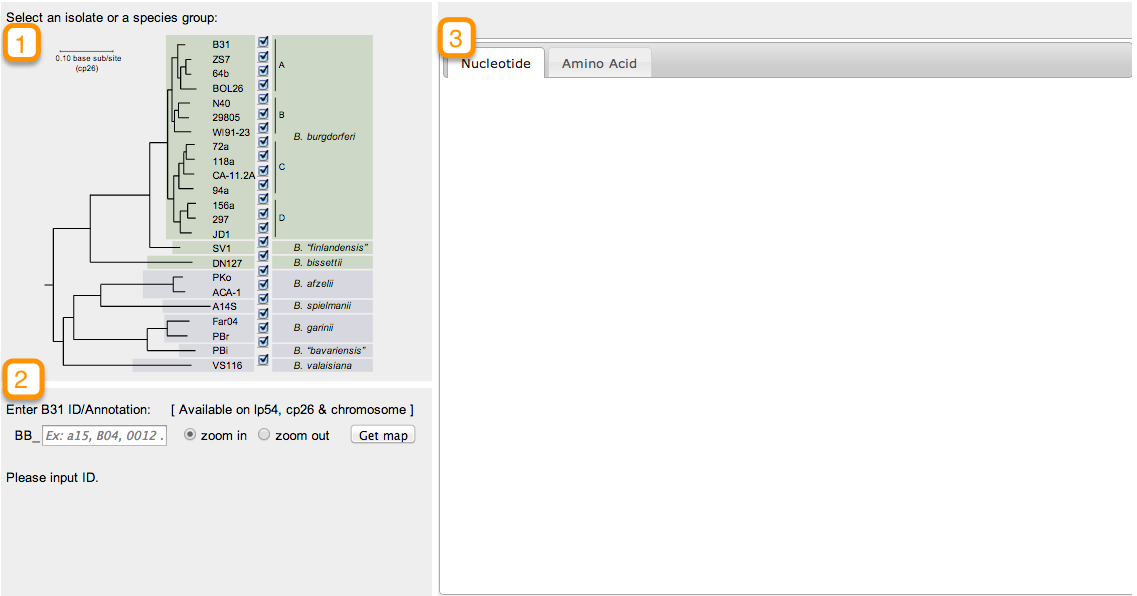
Orthologs are genes that are present in different species that evolved from a common ancestral gene by a speciation event. To retrieve orthologs belonging to the various strains of B. Burgdorferi begin by clicking on the Orthologs tab located at the top of the page:
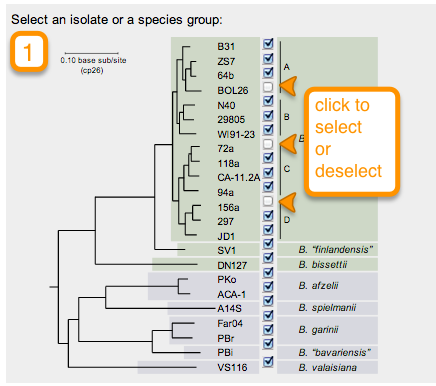
In this new page you will notice a phylogenetic tree of B. Burgdorferi with check boxes to the right of the terminal nodes. Begin by selecting which strains you would like to retrieve information from by checking or unchecking the boxes that correspond to the strain. (Note: by default all strains are selected)
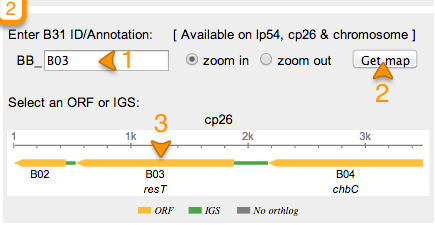
Once you have selected your strain, enter the id or annotation of the ortholog you would like to retrieve.
Ex: B03 -> BB_B03
Once you enter the ID/Annotation (1) click on GET MAP (2). If a result is found, a map will appear with the ortholog of interest as well as flanking orthologs. This map shows a scale bar, flanking orthologs as well as IGS. Each structure is color coded.
Orange -> Open Reading Frame (ORF) Green -> Inter Genetic Sequence (IGS) Gray -> No ortholog
Rectangles are used to depict ORFs with arrow heads indicating direction.
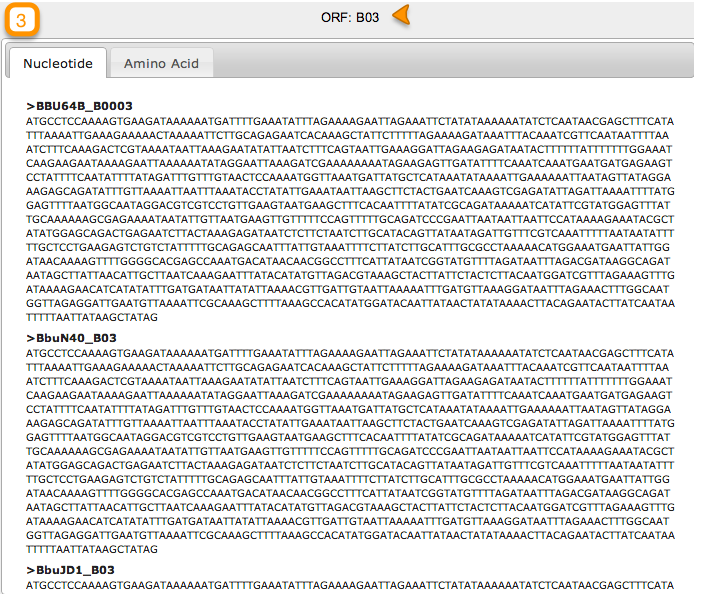
To retrieve sequence information about this ortholog, click on the Orange bar (3 in the above figure).
In the right hand area of your screen, your query results will display. You have the options to retrieve Nucleotide or Amino Acid sequences.
Tutorial 2. How to retrieve orthologous IGS sequences
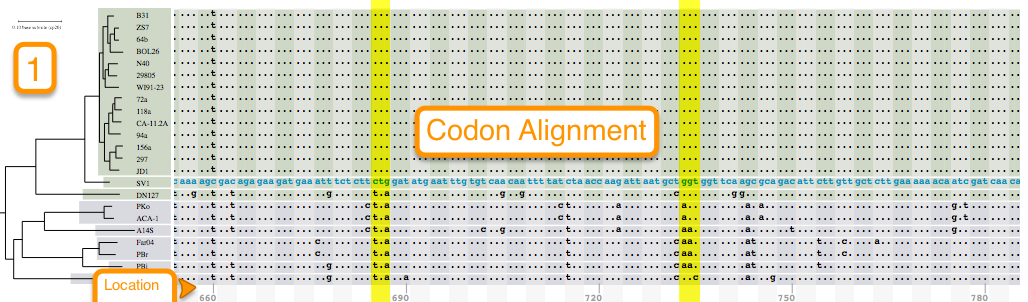
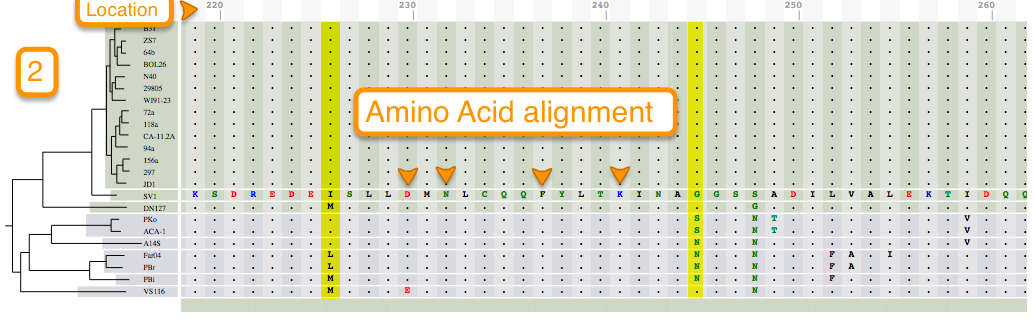
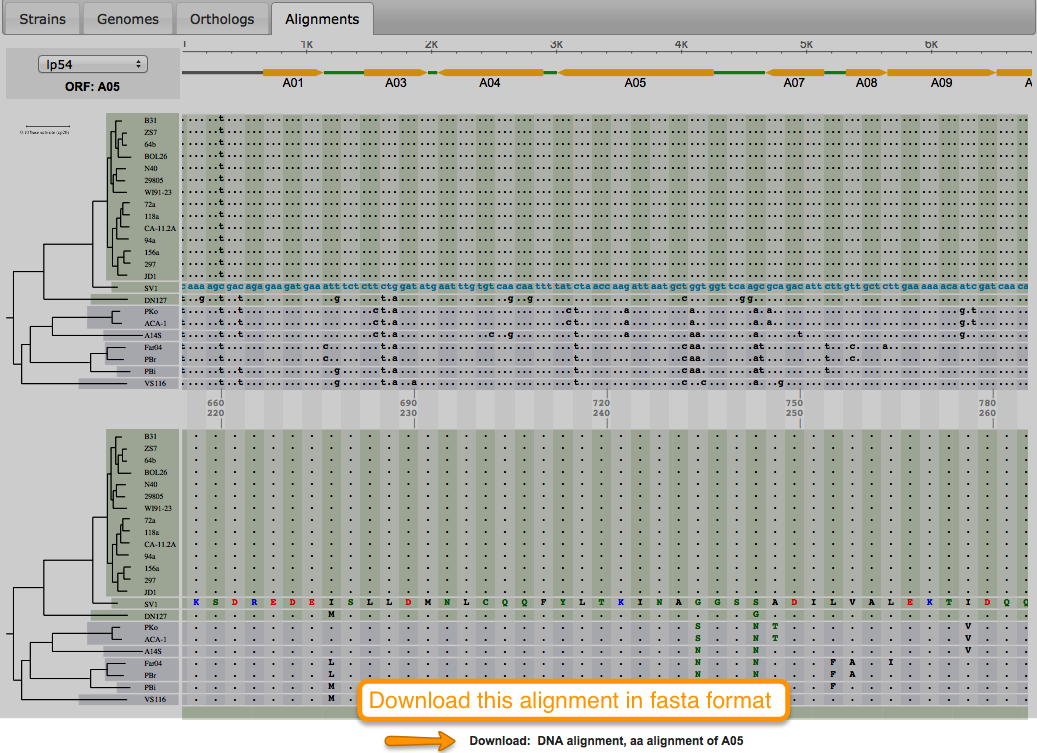
Tutorial 3. How to view codon alignments
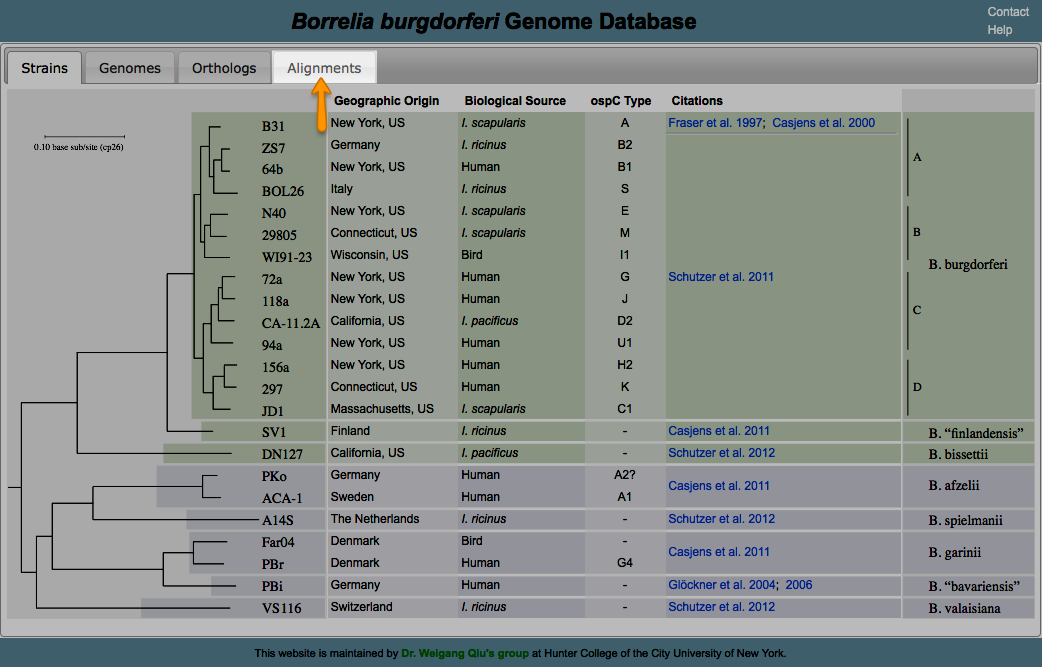
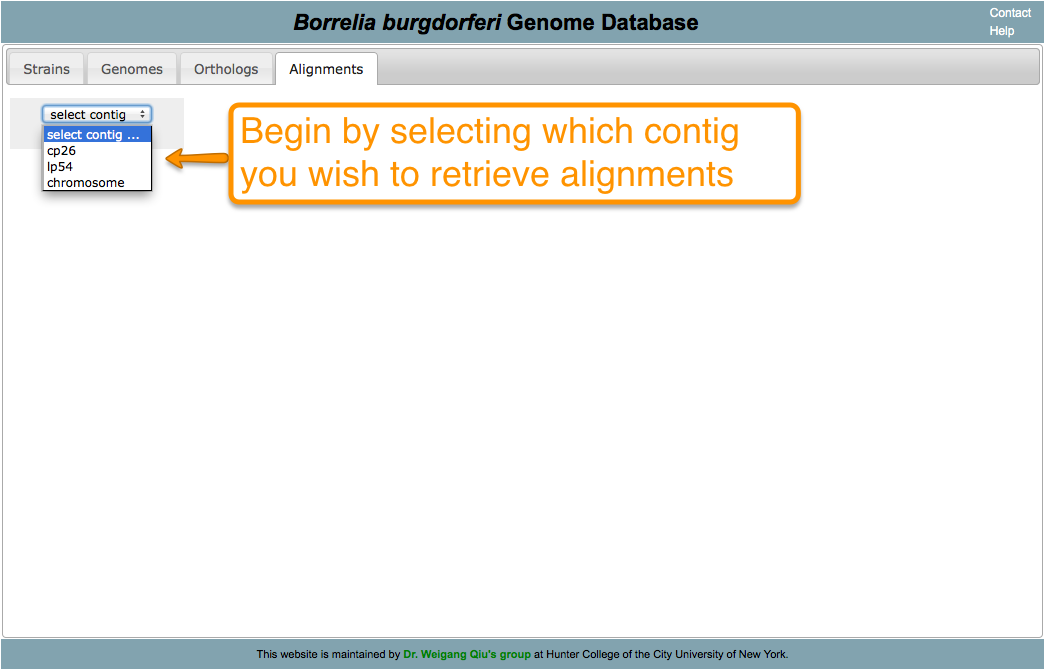
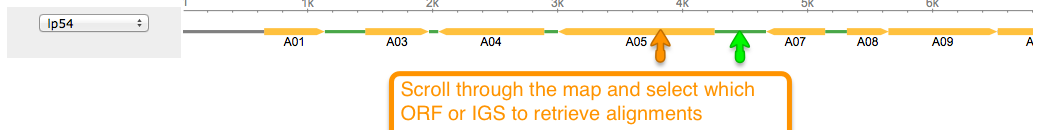
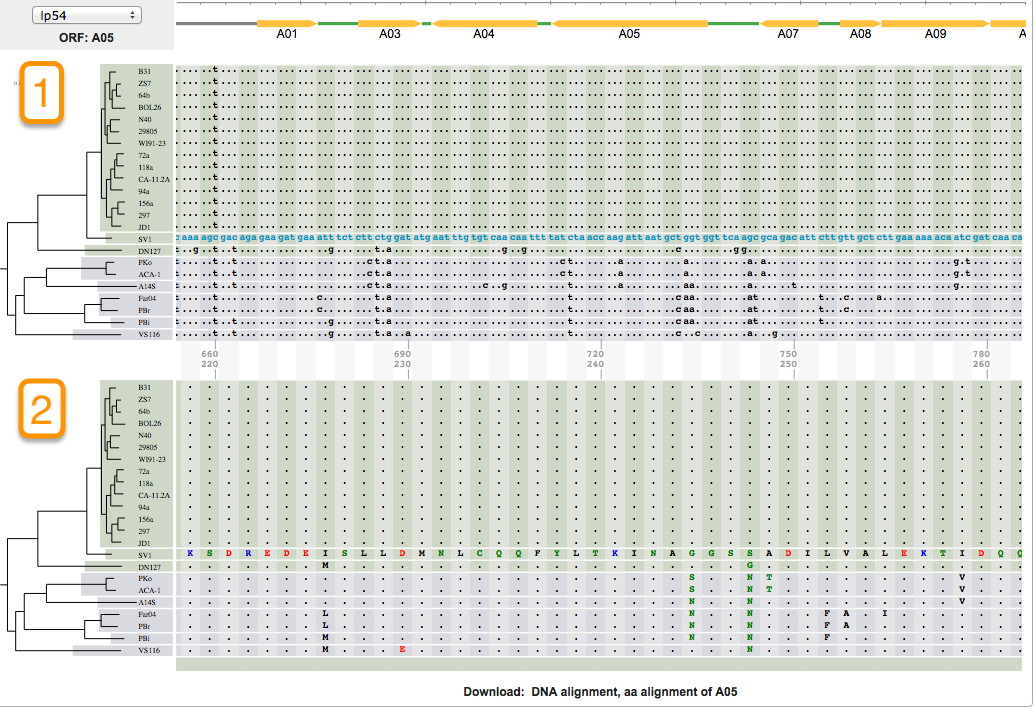
Aligning is a way of arranging sequences (DNA, RNA, or Amino Acid) to identify their similarities. It also helps in inferring functional, structural, and/or evolutionary relationships. One of the features of BorreliaBase is displaying these alignments for you To view alignments for sequences from the Borrelia genome, click on the ALIGNMENTS tab.