BigData 2018
What is evolutionary genomics?
Genomes differ among individuals and change over time. Evolutionary genomics studies genome variability and genome changes based on evolutionary principles. Typical applications include identification of human genome variations associated with diseases and identification of pathogen virulence genes.
Genome changes are studied at two levels: (1) within-species/within-population variations (e.g., human genetic variation), and (2) between-species divergence (e.g., human-mouse comparisons).
The key for analyzing genome variations within species is "population-thinking", the idea that there is no one individual genome that is standard, normal, or disease-free.
The key for comparing genomes across species is "tree-thinking", the idea that evolution happens by diversification (like a branching tree) not by climbing a ladder. There is no such thing as "advanced" or "primitive" species. All living species have the exact same evolutionary distances/time of divergence since the origin of life.
Examples from Qiu Lab
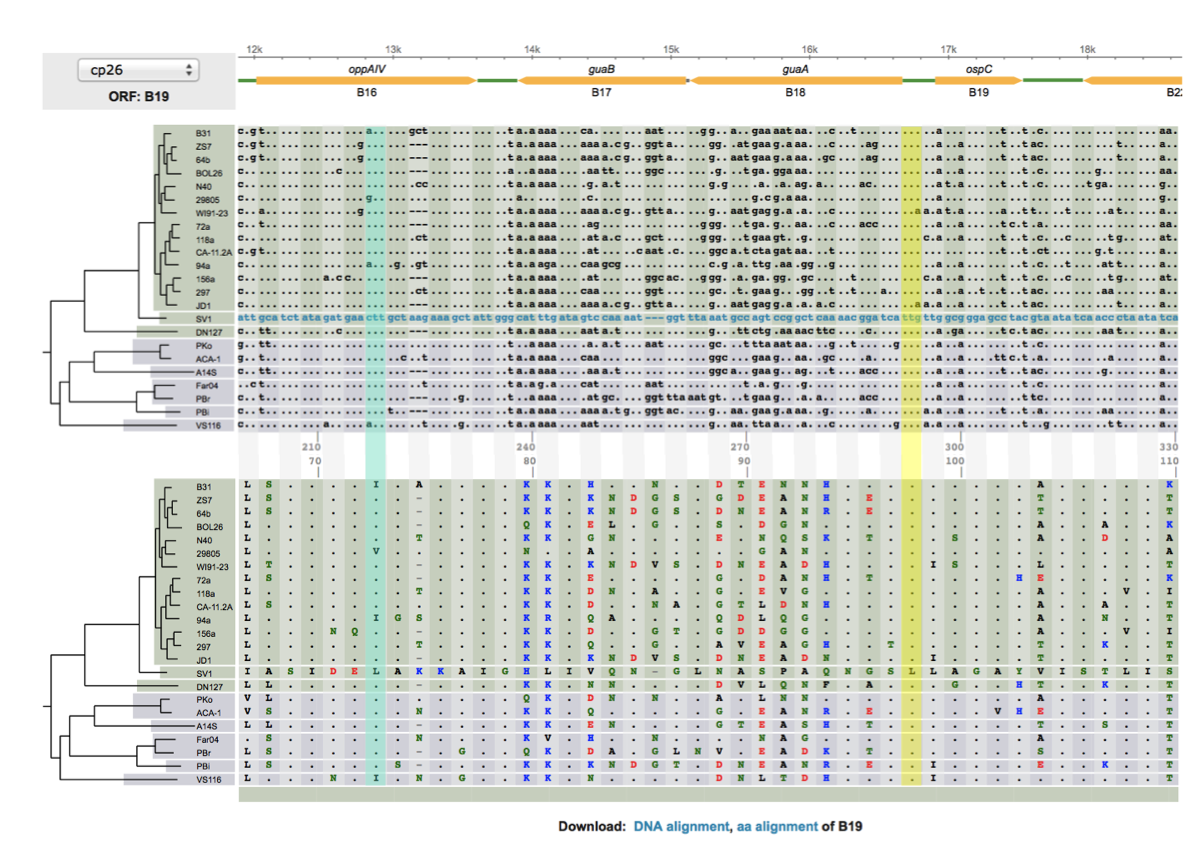
- Comparative genomics of worldwide Lyme disease pathogens
- Evolution of multi-drug antibiotic-resistance Pseudomonas in cancer patients
- Genomic epidemiology of Group B Streptococcus
Reference
- Graur, 2016, Molecular and Genome Evolution, First Edition, Sinauer Associates, Inc. ISBN: 978-1-60535-469-9. Publisher's Website (Student discount: a 15% discount and receive free UPS standard shipping)
- Baum & Smith, 2013. Tree Thinking: an Introduction to Phylogenetic Biology, Roberts & Company Publishers, Inc.
Learning Goals
- Be able to describe evolutionary relationships using phylogenetic trees
- Be able to use command-line tools for batch-processing of genome files