NYRaMP-Informatics-2024
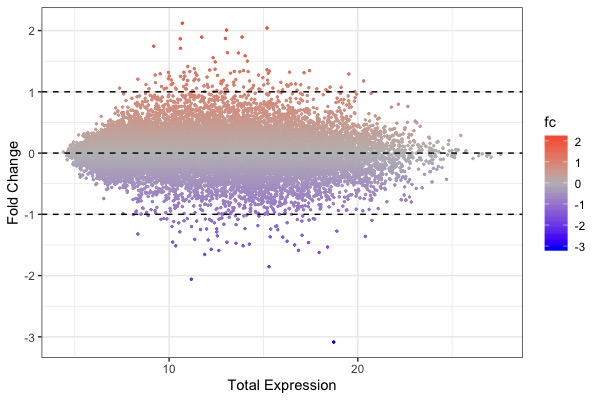
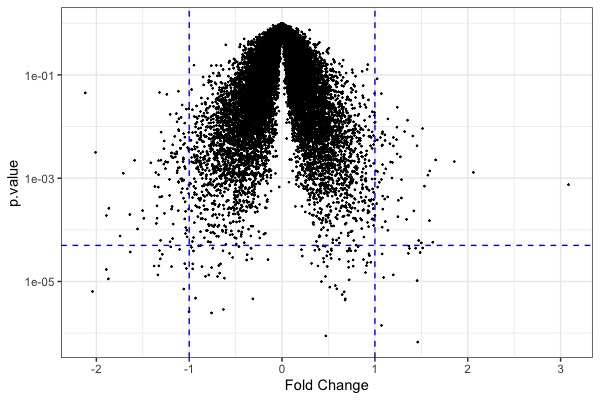
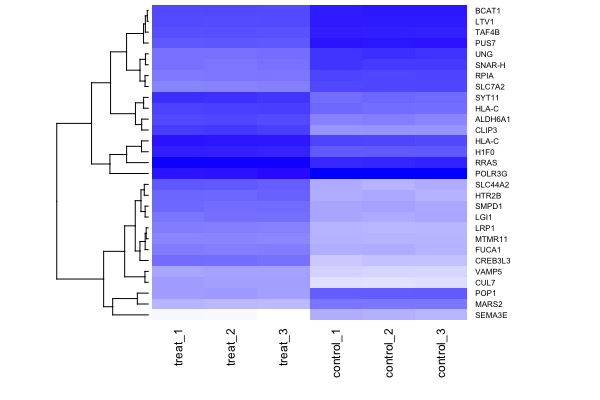
| MA plot | Volcano plot | Heat map |
|---|---|---|
Course Overview
Welcome to Introductory BioMedical Genomics, a seminar course for advanced undergraduates and graduate students. A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and rapid DNA and RNA-sequencing technologies, biomedical sciences are undergoing a rapid & irreversible transformation into a highly data-intensive field, that requires familiarity with concepts in both biology, computational, and data sciences.
Genome information is revolutionizing virtually all aspects of life sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as statistics.
This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises. Students are expected to be able to replicate key results of data analysis from published studies.
The pre-requisites of the course are college-level courses in molecular biology, cell biology, and genetics. Introductory courses in computer programming and statistics are preferred but not strictly required.
Learning goals
By the end of this course successful students will be able to:
- Describe next-generation sequencing (NGS) technologies & contrast it with traditional Sanger sequencing
- Explain applications of NGS technology including pathogen genomics, cancer genomics, human genomic variation, transcriptomics, meta-genomics, epi-genomics, and microbiome.
- Visualize and explore genomics data using R & RStudio
- Replicate key results using a raw data set produced by a primary research paper
Web Links
- Install R base: https://cloud.r-project.org
- Install R Studio (Desktop version): http://www.rstudio.com/download
- Textbook: Introduction to R for Biologists
- Download: R datasets
- A reference book: R for Data Science (Wickharm & Grolemund)