NYRaMP-Informatics-2025
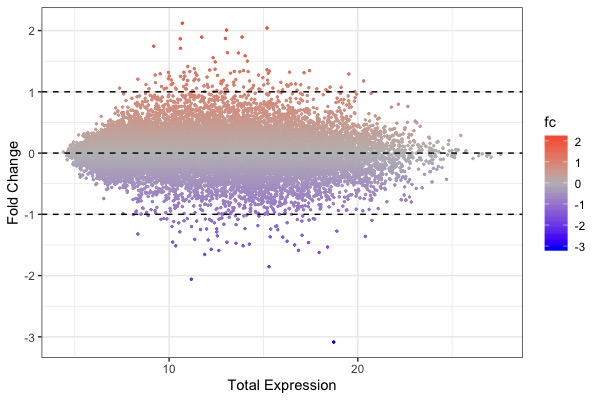
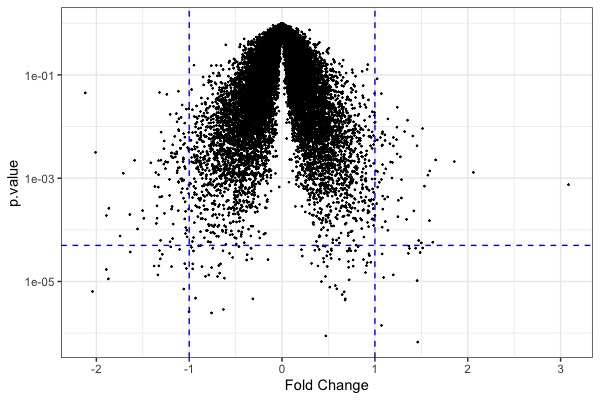
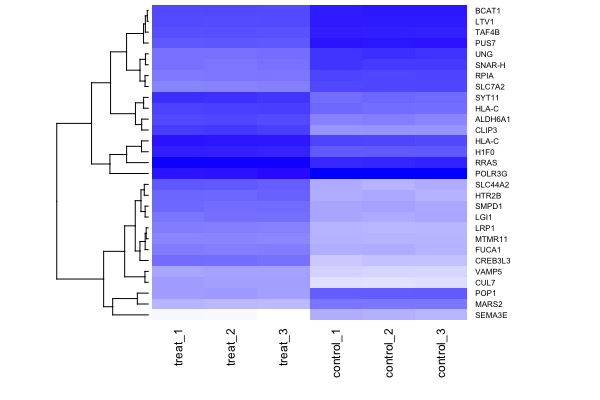
| MA plot | Volcano plot | Heat map |
|---|---|---|
Overview
A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and rapid DNA and RNA-sequencing technologies, biomedical sciences are undergoing a rapid & irreversible transformation into a highly data-intensive field, that requires familiarity with concepts in both biological, computational, and statistical sciences.
Genome information is revolutionizing virtually all aspects of life Sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as statistics.
This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises.
Learning goals
By the end of this workshop students will be able to:
- Manipulate data with R & Rstudio
- Visualize data using R & RStudio
- Analyze microbiome data
Web Links
- Cloud R account (free): https://posit.cloud/; Join the shared work space "NYRaMP-Informatics"
- For your own computer, download the desktop version: https://posit.co/download/rstudio-desktop/
- Textbook: Introduction to R for Biologists
- Download: R datasets
- A reference book: R for Data Science (Wickharm & Grolemund)
Week 1. Aug 5
- Pre-test: visualization, interpretation, and stats. Download file: File:Pre-test.pdf
- Computer/Cloud setup & software download/installation
- R Tutorial 1. Getting started: Basics: interface, packages, variables, objects, functions. Download slides: File:NYRamP bioinformatics 1 slides.pdf
- Session 1 R code: Basic R syntax, working with vectors, and using functions
##### Practice 1 - Together #####
# TASK 1: define a variable that is your name
MyName <- 'Brandon'
print(MyName)
paste('My name is',MyName, sep = ' ')
# TASK 2: output the 3rd and 4th letters in your name using substr function
substr(MyName, start = 3, stop = 4)
# TASK 3: create a vector of the names of all of your Ramp cohort members
roster <- c('Amalya', 'Danny', 'Lorelei', 'Dylan', 'Hadley', 'Brynn', 'Elliot', 'Theo')
# Task 4: check if any of the names have the letters "ic" in them
grepl('an', roster, ignore.case = F)
# Task 5: randomly select 3 names from the roster
sample(roster, size = 3, replace = FALSE)
# TASK 6: combine tasks 4-5
grepl('an', sample(roster, size = 3, replace = F), ignore.case = F)
##### Practice 2 - Independent #####
library(stringr)
# TASK 1: create a character vector for the DNA nucleotides
Nucleotides <- c('A', 'T', 'C', 'G')
# TASK 2: use the "sample" function on your nuc vector to create a DNA sequence of length 200
DNAseq <- sample(Nucleotides, 200, replace = TRUE)
# collapse the vector into a single string
DNAseq2 <- paste(DNAseq, collapse = '')
# TASK 3: Find out if your sequence contains start codons using the "grepl" function; if output = FALSE, start over
grepl('ATG', DNAseq2, ignore.case = F)
# TASK 4: Find all locations of start codons within your sequence using "str_locate_all" function
str_locate_all(string = DNAseq2, pattern = 'ATG')
# TASK 5: use "substring" function to confirm coordinates are actually "ATG"
'ATG' == substr(DNAseq2, start = 96, stop = 98)
# TASK 6: calculate nucleotide % composition using "str_count" function
str_count(string = DNAseq2, pattern = 'A') / 200 * 100
### alternate more advanced way for tasks 5 and 6 ###
coords <- str_locate_all(string = DNAseq2, pattern = 'ATG')
for (i in 1:nrow(coords[[1]])) {
start <- coords[[1]][i, 1]
print('ATG' == substr(DNAseq2, start = start, stop = start+2))
}
for (nuc in Nucleotides) {
print(paste(nuc,' = ', str_count(string = DNAseq2, pattern = nuc)/nchar(DNAseq2)*100, sep = ''))
}
Week 2. Aug 12
- R Tutorial 2. Data manipulation. Download slides: File:R-tutorials-Part-2.pdf
- Practice #2
Week 3. Aug 19
- R Tutorial 3. Data visualization. Lecture slides: File:R-tutorials-Part-3.pdf
- Practice #3
- Demo I. Microbiome data analysis