BigData 2018: Difference between revisions
imported>Weigang mNo edit summary |
imported>Weigang |
||
| Line 21: | Line 21: | ||
* Within-host genome evolution: Evolution of multi-drug antibiotic-resistance Pseudomonas in cancer patients (Figure 2) | * Within-host genome evolution: Evolution of multi-drug antibiotic-resistance Pseudomonas in cancer patients (Figure 2) | ||
==Bioinformatics | ==Bioinformatics Workflow== | ||
* Pathogen isolation -> DNA extraction -> Library preparation -> High-through sequencing | * Pathogen isolation -> DNA extraction -> Library preparation -> High-through sequencing | ||
* De novo genome assembly (canu; velvet; etc) | * De novo genome assembly (canu; velvet; etc) | ||
Revision as of 04:50, 21 June 2018
What is evolutionary genomics?
Genomes differ among individuals and species. Evolutionary genomics studies genome variability and genome changes using evolutionary principles. Typical applications include identification of human genome variations associated with diseases and identification of pathogen virulence genes.
Genome changes are studied at two distinct levels: (1) within-species/within-population variations (e.g., human genetic variation), and (2) between-species divergence (e.g., human-mouse comparisons).
The key for analyzing genome variations within species is "population-thinking", the idea that there is no one individual genome that is standard, normal, or disease-free.
The key for comparing genomes across species is "tree-thinking", the idea that evolution happens by diversification (like a branching tree), not by climbing a ladder. There is no such thing as "advanced" or "primitive" species. All living species have the exact same evolutionary distances/time of divergence since the origin of life.
Case studies from Qiu Lab
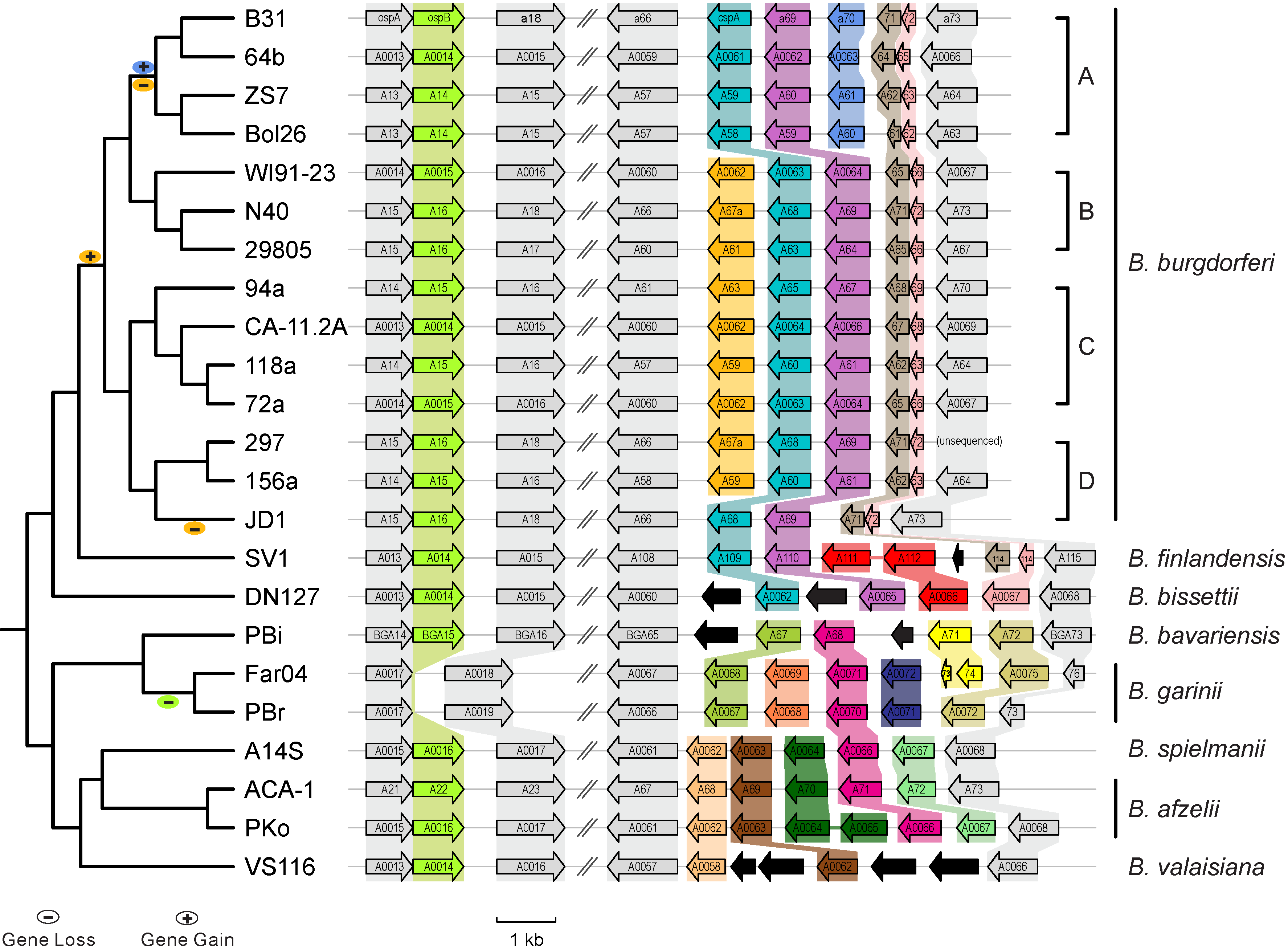
- Between-sepcies genome comparisons: Comparative genomics of worldwide Lyme disease pathogens. BorreliaBase (Figure 1)
- Within-population genome comparison: Genomic epidemiology of Group B Streptococcus: Gene gains & losses associated with Group B Streptococcus virulence
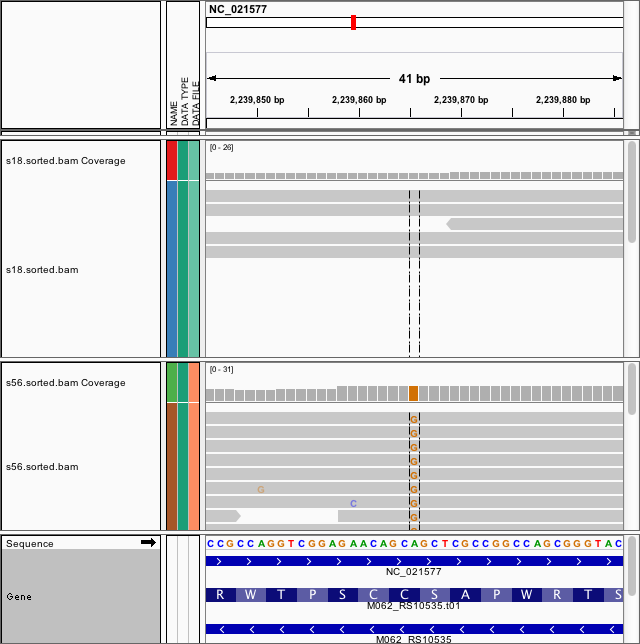
- Within-host genome evolution: Evolution of multi-drug antibiotic-resistance Pseudomonas in cancer patients (Figure 2)
Bioinformatics Workflow
- Pathogen isolation -> DNA extraction -> Library preparation -> High-through sequencing
- De novo genome assembly (canu; velvet; etc)
- Identify reference genome from NCBI database (kraken)
- Variant call (bwa; cortex_var; samtools mpileup)
- Infer genome phylogeny (muscle; reXML)
- Annotation (PATRIC)
- Custom genome browser (JavaScript; D3 library for interactive graphics)
Reference
- Graur, 2016, Molecular and Genome Evolution, First Edition, Sinauer Associates, Inc. ISBN: 978-1-60535-469-9. Publisher's Website
- Baum & Smith, 2013. Tree Thinking: an Introduction to Phylogenetic Biology, Roberts & Company Publishers, Inc.
Learning Goals
- Be able to compare evolutionary relationships using phylogenetic trees
- Be able to use command-line tools for batch-processing of genome files
- Be able to perform genome-wide association analysis on the R platform
Quizzes & Assignments
- "Tree Thinking Quizzes"
- FASTA manipulations with
bioseq