BioMed-R-2021: Difference between revisions
imported>Weigang m (→Jan 30, 2021) |
imported>Weigang m (→May 15, 2020) |
||
| (29 intermediate revisions by the same user not shown) | |||
| Line 30: | Line 30: | ||
* Visualize and explore genomics data using R & RStudio | * Visualize and explore genomics data using R & RStudio | ||
* Replicate key results using a raw data set produced by a primary research paper | * Replicate key results using a raw data set produced by a primary research paper | ||
==A sample of original NGS paper with data sets== | |||
* [https://doi.org/10.1128/mSystems.00939-20 de la Cuesta-Zuluaga J, Spector TD, Youngblut ND, Ley RE. 2021. Genomic insights into adaptations of trimethylamine-utilizing methanogens to diverse habitats, including the human gut. mSystems 6:e00939-20.] | |||
* [https://doi.org/10.1128/mSystems.00033-21 Reiter et al. 2021. Transcriptomics provides a genetic signature of vineyard site and offers insight into vintage-independent inoculated fermentation outcomes. mSystems 6:e00033-21] | |||
* [https://doi.org/10.1128/mSystems.01350-20 Renninger et al (2021). Indoor dust as a matrix for surveillance of COVID-19. mSystems 6:e01350-20.]. [https://datadryad.org/stash/dataset/doi:10.5061/dryad.rn8pk0p8t Data available at Dryad] | |||
* [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-016-1133-7 Litzenburger et al. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol 18, 15 (2017).] | |||
* [https://science.sciencemag.org/content/361/6409/1380 Cao et al. Single-cell chromatin and RNA analysis. Science (2018)] | |||
==Web Links== | ==Web Links== | ||
* Install R base: https://cloud.r-project.org | * Install R base: https://cloud.r-project.org | ||
* Install R Studio (Desktop version): http://www.rstudio.com/download | * Install R Studio (Desktop version): http://www.rstudio.com/download | ||
* R Markdown Template: [http://diverge.hunter.cuny.edu/~weigang/Rmarkdown-template.Rmd R markdown template (by Hector)] | |||
* Textbook: [http://r4all.org/#about Introduction to R for Biologists] | * Textbook: [http://r4all.org/#about Introduction to R for Biologists] | ||
* Download: [http://www.r4all.org/books/datasets R datasets] | * Download: [http://www.r4all.org/books/datasets R datasets] | ||
| Line 73: | Line 81: | ||
===Jan 30, 2021=== | ===Jan 30, 2021=== | ||
* Introduction | * Introduction | ||
* R Tutorial 1: Use interface, basic operations, load data. | * R Tutorial 1: Use interface, basic operations, load data. (slides available on Blackboard) | ||
{| class="wikitable sortable mw-collapsible" | {| class="wikitable sortable mw-collapsible" | ||
! Assignment 1 (15 pts | ! In-class Exercise & Assignment 1 (15 pts) | ||
|- | |- | ||
| | | | ||
* (5 pts) Transform the following "untidy/wide" table into a "tidy/tall" table (print a hard copy) | * (5 pts, Due in-class) Transform the following "untidy/wide" table into a "tidy/tall" table (print a hard copy) | ||
<pre> | <pre> | ||
PropertyName,Density_250m,Density_500m,Density_1000m | PropertyName,Density_250m,Density_500m,Density_1000m | ||
| Line 87: | Line 95: | ||
VanCortlandtPark,0.000550151,0.000979312,0.001372675 | VanCortlandtPark,0.000550151,0.000979312,0.001372675 | ||
</pre> | </pre> | ||
* ( | * (10 pts, Due 2/7) Make a single slide of a primary research paper using next-generation sequencing (NGS) technologies, show the following | ||
** proper citation (authors, title, year, journal, URL) | ** proper citation (authors, title, year, journal, URL) | ||
** NGS method (Illumina, PacBio, or NanoPore) | ** NGS method (Illumina, PacBio, or NanoPore) | ||
| Line 94: | Line 102: | ||
** raw data table (show first few columns and first few rows) | ** raw data table (show first few columns and first few rows) | ||
** for example, a student has worked on tissue regeneration, the search in PubMed with key words "regeneration zebra fish transcriptome" found the following primary paper as the best because of the high quality of journal and the availability of raw data: https://www.ncbi.nlm.nih.gov/pubmed/28096348 | ** for example, a student has worked on tissue regeneration, the search in PubMed with key words "regeneration zebra fish transcriptome" found the following primary paper as the best because of the high quality of journal and the availability of raw data: https://www.ncbi.nlm.nih.gov/pubmed/28096348 | ||
** Papers to AVOID: | |||
*** Reviews (not original research paper): e.g.,[https://pubmed.ncbi.nlm.nih.gov/30089861/ a review paper (usually appear first on PubMed; SKIP)] | |||
*** Original research, but no computer readable raw data: e.g., [https://www-ncbi-nlm-nih-gov.proxy.wexler.hunter.cuny.edu/pmc/articles/PMC7737787/ Raw data not readable (PDF is not computer-readable)] | |||
|} | |} | ||
===Feb 6, 2021=== | ===Feb 6, 2021=== | ||
* Introduction to NGS: | * Introduction to NGS: (slides available on Blackboard) | ||
* 1-slide presentations on Next-Generation Sequencing Technologies (Group I) | * 1-slide presentations on Next-Generation Sequencing Technologies (Group I) | ||
* R Tutorial, Part 2. Data manipulation with dplyr. Slides: [[File:R-tutorials-2.pdf|thumbnail]] | * R Tutorial, Part 2. Data manipulation with dplyr. Slides: [[File:R-tutorials-2.pdf|thumbnail]] | ||
{| class="wikitable sortable mw-collapsible" | {| class="wikitable sortable mw-collapsible" | ||
! | ! In-class Exercise 2 (10 pts; Due in class) | ||
|- | |- | ||
| | | | ||
* | * Show R commands for the following operations | ||
* | ** load the "tidyverse" library | ||
** load the "iris" data | |||
** Select columns "Sepal.Length" & "Species" | ** Select columns "Sepal.Length" & "Species" | ||
** Filter rows 2 through 10 | ** Filter rows 2 through 10 | ||
** Add a column "logSepalLength" by taking the logarithm of the said column | ** Add a column "logSepalLength" by taking the logarithm of the said column | ||
** Calculate mean and standard deviation of Petal.Length in each species | ** Calculate mean and standard deviation of Petal.Length in each species | ||
* | ** Save all commands in a script "in-class-ex-2.R" | ||
|} | |} | ||
* Assignment 1 Due next day | |||
===Feb 13, 2021=== | ===Feb 13, 2021=== | ||
* NGS presentations | * NGS presentations | ||
* R Tutorial. Part 3. Data visualization with ggplot2. Slides: [[File:R-tutorials-3.pdf|thumbnail]] | * R Tutorial. Part 3. Data visualization with ggplot2. Slides: [[File:R-tutorials-3.pdf|thumbnail]] | ||
* | * Assignment 2: Submit R-Markdown with scatter plot & boxplot | ||
===Feb 20, 2020=== | ===Feb 20, 2020=== | ||
* R Tutorial: Part 4. BioStat (chi-square & t-test) Lecture slides: [[File:R-tutorial-4.pdf|thumbnail]] | * R Tutorial: Part 4. BioStat (chi-square & t-test) Lecture slides: [[File:R-tutorial-4.pdf|thumbnail]] | ||
* Presentation | |||
* Assignment 3 (10 pts). R Markdown upload (by Wed 8pm) | |||
===Feb 27, 2020=== | ===Feb 27, 2020=== | ||
* | * Review | ||
* | * Quiz (30 pts) | ||
** NGS slides & R tutorial Slides | |||
** Excluding the four statistical tests | |||
** Openbook but due in class (on Blackboard) | |||
===March 6, 2020=== | ===March 6, 2020=== | ||
* | * R Tutorial: Part 4. BioStat (regression & ANOVA) [[File:R-tutorial-5.pdf|thumbnail]] | ||
* | * Paper review | ||
* Assignment 4 (10 pts). Submit R markdown on Blackboard | |||
===March 13, 2020=== | ===March 13, 2020=== | ||
* | * Paper review | ||
* Review for mid-term exam: 6 PDF presentations (intro to NGS & 5 R-tutorials) | |||
===March 20, 2020=== | ===March 20, 2020=== | ||
* | * Mid-term | ||
* | ** Focus on four statistical tests (with visualization | ||
* | ** Submit sectioned R markdown PDF: [http://diverge.hunter.cuny.edu/~weigang/Rmarkdown-template.Rmd R markdown template (by Hector)] | ||
* R | |||
===March 27, 2020=== | ===March 27, 2020=== | ||
| Line 159: | Line 161: | ||
===April 3, 2020=== | ===April 3, 2020=== | ||
* | * No class: Holiday break | ||
===April 10, 2020=== | ===April 10, 2020=== | ||
* | * Paper assignments & break into project groups | ||
* | * For final presentation (10 pts) | ||
** Each member will present his or her part | |||
* For final report (90), you are required to: | |||
** Read the paper and identify a dataset to replicate | |||
** Create an R markdown file to record your work | |||
** Produce a final WORD or PDF file as final report | |||
* Case study 1: Restriction/Modification system in Lyme pathogen: [https://jb.asm.org/content/200/24/e00395-18 Casselii et al (2018)] | |||
===April 17, 2020=== | ===April 17, 2020=== | ||
* | * Group presentation #1. Summary slides | ||
===April 24, 2020=== | ===April 24, 2020=== | ||
* | * Group presentation #2. R Markdown & Data import | ||
* | * I will hear group presentation Round #2. I will grade individual performance by the following rubric: | ||
* | ** Biologist: Be prepared to answer my questions regarding the study background, question, and significance | ||
** Writer: Be prepared to show R Markdown (in R Studio) that included ALL sections from the last week's slide | |||
** Data Scientist: Be prepared to show data table that has been read into R Studio & show a preliminary graph & visualization | |||
** Statistician: Be prepared to answer questions on what is the statistical null hypothesis and what test to perform | |||
===May 1, 2020=== | ===May 1, 2020=== | ||
* | * Group presentation #3. R Markdown & Data analysis | ||
===May 8, 2020=== | ===May 8, 2020=== | ||
* Consultation (no | * Consultation by appointment (no live session) | ||
===May 15, 2020=== | ===May 15, 2020=== | ||
* Consultation (no | * Consultation by appointment (no live session) | ||
* '''Submit your Teacher's Evaluation''', using either: | |||
** Personal computer at [http://www.hunter.cuny.edu/te www.hunter.cuny.edu/te]; or, | |||
** Smartphone at [http://www.hunter.cuny.edu/mobilete www.hunter.cuny.edu/mobilete] | |||
===May 22, 2020=== | ===May 22, 2020=== | ||
* Friday, 5pm: Final report Due (Blackboard submission) | * Friday, 5pm: Final report Due (Blackboard submission) | ||
* '''Your final report (100 pts) should include the following required components''': | |||
** (10 pts) Section 1. Background & Objectives. Describe (a) the overall goal of the study; (b) the specific question to be addressed by your dataset | |||
** (20 pts) Section 2. Material & Methods. Describe experimental design, i.e., how your assigned data set was generated, including the nature of the biological samples, sample size, number of replicates (biological & technical), controls (if any), sequencing technologies. Hint: Fig S1 | |||
** (40 pts) Section 3. R codes & graphs. Show R codes with comments for individual commands. Graphics should be as close to the published figure as possible (e.g., with proper axis labels) | |||
** (10 pts) Section 4. Statistical analysis. Show mull hypothesis and p-value. Draw statistical conclusion | |||
** (5 pts) Section 5. Conclusion. Draw biological conclusions of your analysis | |||
** (5 pts) Section 6. Citations/source/URL to paper, your dataset, and methods | |||
Latest revision as of 02:12, 2 May 2021
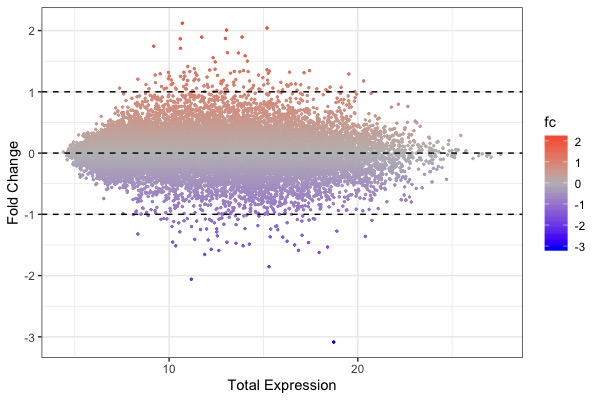
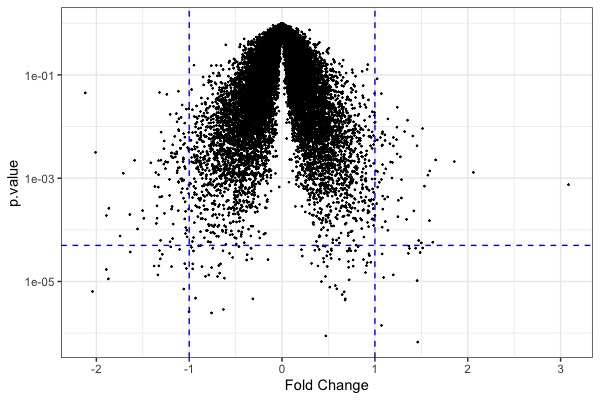
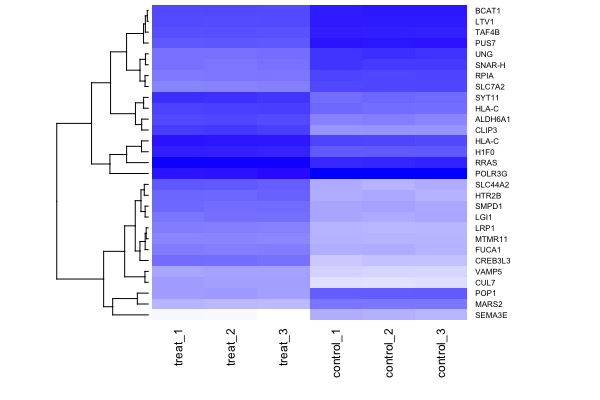
| MA plot | Volcano plot | Heat map |
|---|---|---|
Course Overview
Welcome to Introductory BioMedical Genomics, a seminar course for advanced undergraduates and graduate students. A genome is the total genetic content of an organism. Driven by breakthroughs such as the decoding of the first human genome and rapid DNA and RNA-sequencing technologies, biomedical sciences are undergoing a rapid & irreversible transformation into a highly data-intensive field, that requires familiarity with concepts in both biology, computational, and data sciences.
Genome information is revolutionizing virtually all aspects of life sciences including basic research, medicine, and agriculture. Meanwhile, use of genomic data requires life scientists to be familiar with concepts and skills in biology, computer science, as well as statistics.
This workshop is designed to introduce computational analysis of genomic data through hands-on computational exercises. Students are expected to be able to replicate key results of data analysis from published studies.
The pre-requisites of the course are college-level courses in molecular biology, cell biology, and genetics. Introductory courses in computer programming and statistics are preferred but not strictly required.
Learning goals
By the end of this course successful students will be able to:
- Describe next-generation sequencing (NGS) technologies & contrast it with traditional Sanger sequencing
- Explain applications of NGS technology including pathogen genomics, cancer genomics, human genomic variation, transcriptomics, meta-genomics, epi-genomics, and microbiome.
- Visualize and explore genomics data using R & RStudio
- Replicate key results using a raw data set produced by a primary research paper
A sample of original NGS paper with data sets
- de la Cuesta-Zuluaga J, Spector TD, Youngblut ND, Ley RE. 2021. Genomic insights into adaptations of trimethylamine-utilizing methanogens to diverse habitats, including the human gut. mSystems 6:e00939-20.
- Reiter et al. 2021. Transcriptomics provides a genetic signature of vineyard site and offers insight into vintage-independent inoculated fermentation outcomes. mSystems 6:e00033-21
- Renninger et al (2021). Indoor dust as a matrix for surveillance of COVID-19. mSystems 6:e01350-20.. Data available at Dryad
- Litzenburger et al. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol 18, 15 (2017).
- Cao et al. Single-cell chromatin and RNA analysis. Science (2018)
Web Links
- Install R base: https://cloud.r-project.org
- Install R Studio (Desktop version): http://www.rstudio.com/download
- R Markdown Template: R markdown template (by Hector)
- Textbook: Introduction to R for Biologists
- Download: R datasets
- A reference book: R for Data Science (Wickharm & Grolemund)
Quizzes and Exams
Student performance will be evaluated by attendance, weekly assignments, quizzes, and a final report in R Markdown:
- Attendance & In-class participation: 100 pts
- Assignments: 5 x 10 = 50 pts
- Quizzes: 2 x 25 pts = 50 pts
- Mid-term: 50 pts
- Final presentation & report: 50 pts
Total: 300 pts
Tips for Success
To maximize the your experience we strongly recommend the following strategies:
- Follow the directions for efficiently, finding high-impact papers, reading science research papers and preparing presentations.
- Read the papers, watch required videos and do the exercises regularly, long before you attend class.
- Attend all classes, as required. Late arrival results in loss of points.
- Keep up with online exercises. Don’t wait until the due date to start tasks.
- Take notes or annotate slides while attending the lectures.
- Listen actively and participate in class and in online discussions.
- Review and summarize material within 24 hrs after class.
- Observe the deadlines for submitting your work. Late submissions incur penalties.
- Put away cell phones, do not TM, email or play computer games in class.
Hunter/CUNY Policies
- Policy on Academic Integrity
Hunter College regards acts of academic dishonesty (e.g., plagiarism, cheating on homework, online exercises or examinations, obtaining unfair advantage, and falsification of records and official documents) as serious offenses against the values of intellectual honesty. The College is committed to enforcing the CUNY Policy on Academic Integrity, and we will pursue cases of academic dishonesty according to the Hunter College Academic Integrity Procedures. Students will be asked to read this statement before exams.
- ADA Policy
In compliance with the American Disability Act of 1990 (ADA) and with Section 504 of the Rehabilitation Act of 1973, Hunter College is committed to ensuring educational parity and accommodations for all students with documented disabilities and/or medical conditions. It is recommended that all students with documented disabilities (Emotional, Medical, Physical, and/or Learning) consult the Office of AccessABILITY, located in Room E1214B, to secure necessary academic accommodations. For further information and assistance, please call: (212) 772- 4857 or (212) 650-3230.
- Syllabus Policy
Except for changes that substantially affect implementation of the evaluation (grading) statement, this syllabus is a guide for the course and is subject to change with advance notice, announced in class or posted on Blackboard.
Course Schedule
Jan 30, 2021
- Introduction
- R Tutorial 1: Use interface, basic operations, load data. (slides available on Blackboard)
| In-class Exercise & Assignment 1 (15 pts) |
|---|
PropertyName,Density_250m,Density_500m,Density_1000m HighbridgePark,0.006561319,0.009462031,0.010578611 BronxRiverParkway,0.001318749,0.001978858,0.002652118 CrotonaPark,0.009412087,0.01164712,0.01202321 ClaremontPark,0.016391948,0.019972485,0.020350481 VanCortlandtPark,0.000550151,0.000979312,0.001372675
|
Feb 6, 2021
- Introduction to NGS: (slides available on Blackboard)
- 1-slide presentations on Next-Generation Sequencing Technologies (Group I)
- R Tutorial, Part 2. Data manipulation with dplyr. Slides:
| In-class Exercise 2 (10 pts; Due in class) |
|---|
|
- Assignment 1 Due next day
Feb 13, 2021
- NGS presentations
- R Tutorial. Part 3. Data visualization with ggplot2. Slides:
- Assignment 2: Submit R-Markdown with scatter plot & boxplot
Feb 20, 2020
- R Tutorial: Part 4. BioStat (chi-square & t-test) Lecture slides:
- Presentation
- Assignment 3 (10 pts). R Markdown upload (by Wed 8pm)
Feb 27, 2020
- Review
- Quiz (30 pts)
- NGS slides & R tutorial Slides
- Excluding the four statistical tests
- Openbook but due in class (on Blackboard)
March 6, 2020
- R Tutorial: Part 4. BioStat (regression & ANOVA)
- Paper review
- Assignment 4 (10 pts). Submit R markdown on Blackboard
March 13, 2020
- Paper review
- Review for mid-term exam: 6 PDF presentations (intro to NGS & 5 R-tutorials)
March 20, 2020
- Mid-term
- Focus on four statistical tests (with visualization
- Submit sectioned R markdown PDF: R markdown template (by Hector)
March 27, 2020
- No class: Spring Break
April 3, 2020
- No class: Holiday break
April 10, 2020
- Paper assignments & break into project groups
- For final presentation (10 pts)
- Each member will present his or her part
- For final report (90), you are required to:
- Read the paper and identify a dataset to replicate
- Create an R markdown file to record your work
- Produce a final WORD or PDF file as final report
- Case study 1: Restriction/Modification system in Lyme pathogen: Casselii et al (2018)
April 17, 2020
- Group presentation #1. Summary slides
April 24, 2020
- Group presentation #2. R Markdown & Data import
- I will hear group presentation Round #2. I will grade individual performance by the following rubric:
- Biologist: Be prepared to answer my questions regarding the study background, question, and significance
- Writer: Be prepared to show R Markdown (in R Studio) that included ALL sections from the last week's slide
- Data Scientist: Be prepared to show data table that has been read into R Studio & show a preliminary graph & visualization
- Statistician: Be prepared to answer questions on what is the statistical null hypothesis and what test to perform
May 1, 2020
- Group presentation #3. R Markdown & Data analysis
May 8, 2020
- Consultation by appointment (no live session)
May 15, 2020
- Consultation by appointment (no live session)
- Submit your Teacher's Evaluation, using either:
- Personal computer at www.hunter.cuny.edu/te; or,
- Smartphone at www.hunter.cuny.edu/mobilete
May 22, 2020
- Friday, 5pm: Final report Due (Blackboard submission)
- Your final report (100 pts) should include the following required components:
- (10 pts) Section 1. Background & Objectives. Describe (a) the overall goal of the study; (b) the specific question to be addressed by your dataset
- (20 pts) Section 2. Material & Methods. Describe experimental design, i.e., how your assigned data set was generated, including the nature of the biological samples, sample size, number of replicates (biological & technical), controls (if any), sequencing technologies. Hint: Fig S1
- (40 pts) Section 3. R codes & graphs. Show R codes with comments for individual commands. Graphics should be as close to the published figure as possible (e.g., with proper axis labels)
- (10 pts) Section 4. Statistical analysis. Show mull hypothesis and p-value. Draw statistical conclusion
- (5 pts) Section 5. Conclusion. Draw biological conclusions of your analysis
- (5 pts) Section 6. Citations/source/URL to paper, your dataset, and methods