BorreliaBase Help: Difference between revisions
imported>Girish |
imported>Girish |
||
| Line 72: | Line 72: | ||
[[File:Tut1-1.png|400px]] | [[File:Tut1-1.png|400px]] | ||
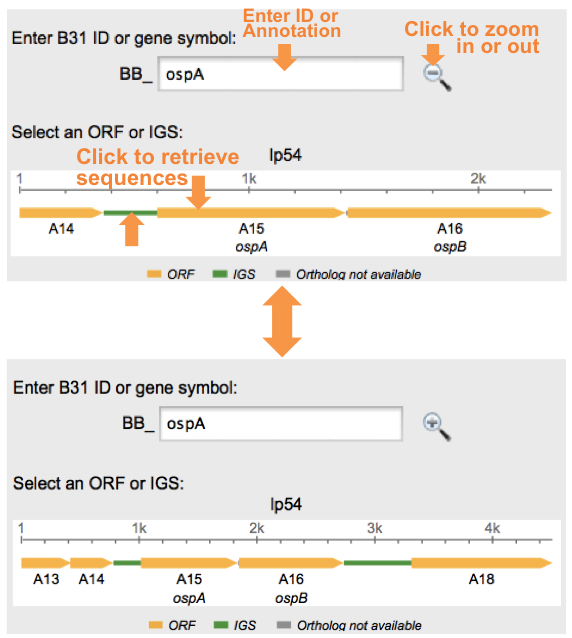
Intergeneic regions will appear as green rectangles. Enter the ID or annotation of a gene, this will generate a map of flanking ORFS in addition to the IGS that separate them. By using the magnifying glass, additional ORFS and IGS can be seen. | |||
* Orange -> Open Reading Frame (ORF) | * Orange -> Open Reading Frame (ORF) | ||
| Line 79: | Line 80: | ||
[[File:Tut2-2.png|400px]] | [[File:Tut2-2.png|400px]] | ||
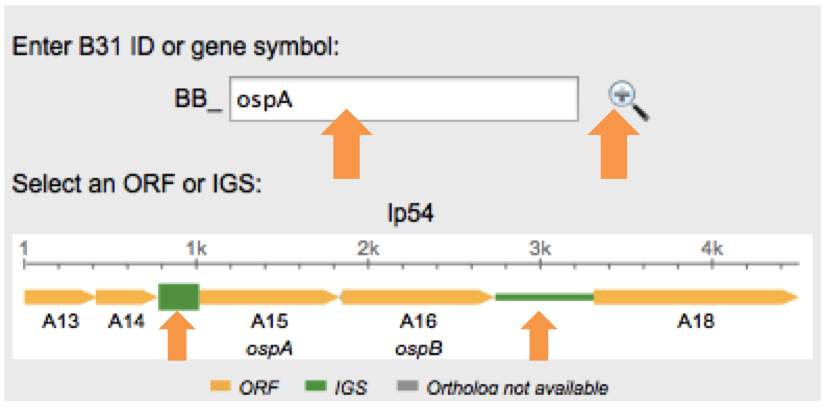
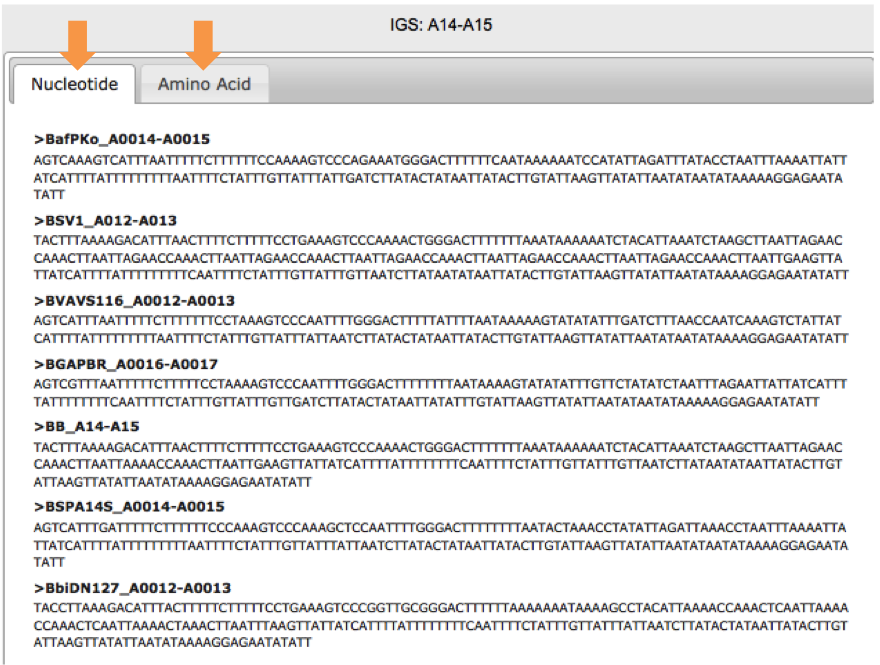
Finally select the IGS by clicking on the corresponding green block. | |||
[[File:Tut2-3.png|400px]] | [[File:Tut2-3.png|400px]] | ||
Revision as of 01:31, 2 January 2014
What is BorreliaBase?
BorreliaBase focuses on providing comparative information among sequenced Borrelia genomes. Current comparative-genomics features include orthologous ORF and IGS (intergenic-spacer) sequences on the main chromosome and the lp54, and cp26 plasmids.
At the center of BorreliaBase is a phylogeny based on genome-wide SNPs. This phylogeny is used as a guide for selecting and de-selecting genomes to be visualized and, more importantly, for providing evolutionary expectations on sequence variability at each genomic locus. Functional insights of an ORF or IGS could be gained by searching for departure from evolutionary expectations, such as the greatly elevated sequence variability at ospC, reduced sequence variability at the promoter region of the ospA/B operon, and a cluster of homoplasies at B22 caused by cross-species recombination.
How often is BorreliaBase updated?
Data in BorreliaBase are manually curated. Updates occur continuous as more orthologs are identified, problematic members of orthologs are removed, ORF sequences are re-annotated (for start-codon positions), and more genomes are added.
Who is behind BorreliaBase?
BorreliaBase is supported by researchers and physicians from the following institutions:
- Dr Weigang Qiu, Department of Biological Sciences and the Center for Gene Structure and Function, Hunter College, City University of New York, New York, New York 10065
- Drs Emmanuel Mongodin and Claire Fraser, Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, Maryland 21201
- Dr Sherwood Casjens, Department of Pathology, University of Utah School of Medicine, Salt Lake City, Utah 84112
- Dr Steven Schutzer, Department of Medicine, University of Medicine and Dentistry of New Jersey-New Jersey Medical School, Newark, New Jersey 07103
- Dr Ben Luft, Department of Medicine, Health Science Center, Stony Brook University, Stony Brook, New York 11794
How is BorreliaBase supported?
BorreliaBase is currently supported by a Public Health Service grant (AI107955) from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH). However, the content of BorreliaBase is solely the responsibility of the research group (see above) and does not represent the official endorsement of NIAID or NIH.
How to send comments and suggestions
Please email Dr Weigang Qiu (weigang at genectr.hunter.cuny.edu)'s group at Hunter College, City University of New York. We welcome your feedback.
How to Cite BorreliaBase?
- To cite the database itself:
- Di et al (2014). In preparation
- To cite the genome phylogeny and annotation:
- Mongodin EF, Casjens SR, Bruno JF, Xu Y, Drabek EF, Riley DR, Cantarel BL, Pagan PE, Hernandez YA, Vargas LC, Dunn JJ, Schutzer SE, Fraser CM, Qiu WG, Luft BJ. (2013). Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics 14(1):693. Free Full Text
- To cite orthologous ORF sequences:
- Haven J, Vargas LC, Mongodin EF, Xue V, Hernandez Y, Pagan P, Fraser-Liggett CM, Schutzer SE, Luft BJ, Casjens SR, Qiu WG. (2011). Pervasive Recombination and Sympatric Genome Diversification Driven by Frequency-Dependent Selection in Borrelia burgdorferi, the Lyme disease Bacterium. Genetics. 189:951-966. Free full text
- To cite orthologous IGS sequences:
- Martin et al (2014). In preparation
Tutorial 1. How to retrieve orthologous ORF sequences?
Orthologs are genes that are present in different species that evolved from a common ancestral gene by a speciation event. To retrieve orthologs belonging to the various strains of B. Burgdorferi begin by clicking on the Orthologs tab located at the top of the page:
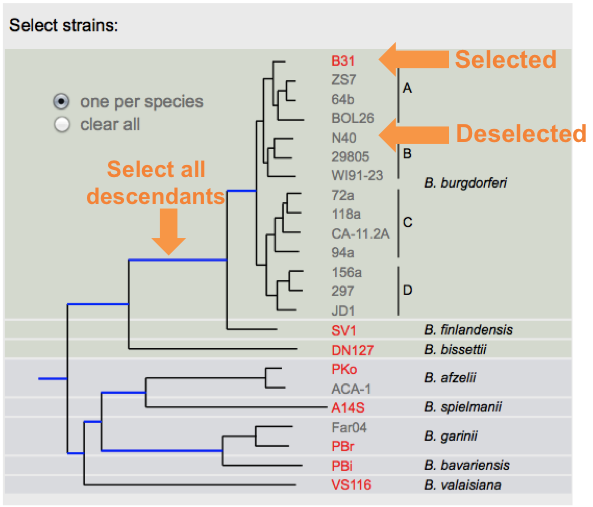
In this new page you will notice a phylogenetic tree of B. Burgdorferi with check boxes to the right of the terminal nodes. Begin by selecting which strains you would like to retrieve information from by clicking the id of the strain, alternatively you can select one strain per species (buttons) or by clicking on a blue branch to select all descendants:
Once you have selected your strain, enter the id or annotation of the ortholog you would like to retrieve.
Ex: B03 -> BB_B03
Once you enter the ID/Annotation, press enter. If a result is found, a map will appear with the ortholog of interest as well as flanking orthologs. This map shows a scale bar, flanking orthologs as well as IGS. Each structure is color coded.
- Orange -> Open Reading Frame (ORF)
- Green -> Inter Genetic Sequence (IGS)
- Gray -> No Information
Rectangles are used to depict ORFs with arrow heads indicating direction.
To retrieve sequence information about this ortholog, click on the Orange bar (3 in the above figure).
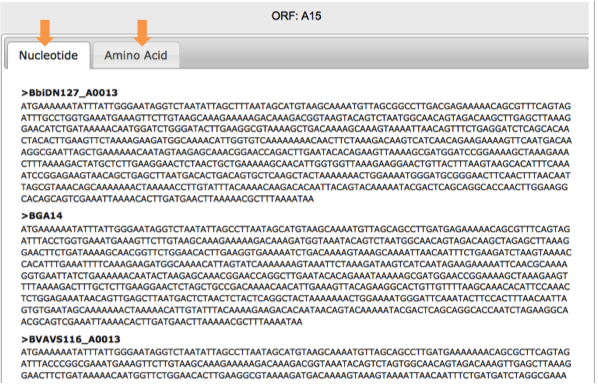
In the right hand area of your screen, your query results will display. You have the options to retrieve Nucleotide or Amino Acid sequences.
Tutorial 2. How to retrieve orthologous IGS sequences
The sequences between genes has been thought to have some regulatory function. Here we catalog conserved putative functional IGS within the core genome of B. burgdorferi. Specific IGS can be retrieved through the ORTHOLOGS Tab.
Select which species to retrieve the IGS from by using the phylogenetic tree as a guide. Selected strains will appear in red bolded font. Entire lineages can be selected by clicking on the blue shaded branches.
Intergeneic regions will appear as green rectangles. Enter the ID or annotation of a gene, this will generate a map of flanking ORFS in addition to the IGS that separate them. By using the magnifying glass, additional ORFS and IGS can be seen.
- Orange -> Open Reading Frame (ORF)
- Green -> Inter Genetic Sequence (IGS)
- Gray -> No Information
Finally select the IGS by clicking on the corresponding green block.
Tutorial 3. How to view codon alignments
Aligning is a way of arranging sequences (DNA, RNA, or Amino Acid) to identify their similarities. It also helps in inferring functional, structural, and/or evolutionary relationships. One of the features of BorreliaBase is displaying these alignments for you To view alignments for sequences from the Borrelia genome, click on the ALIGNMENTS tab.
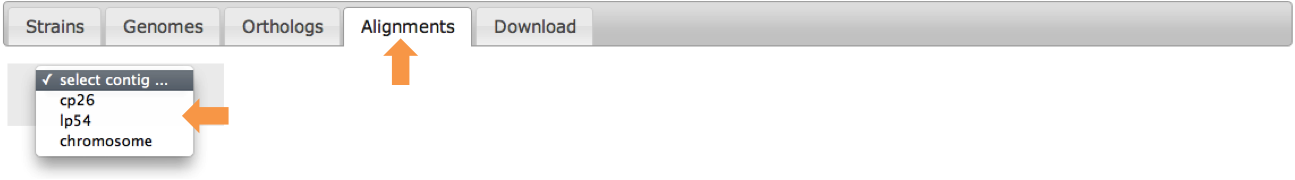
Begin by selecting from which contig of the core genome you would like to retrieve alignment information.
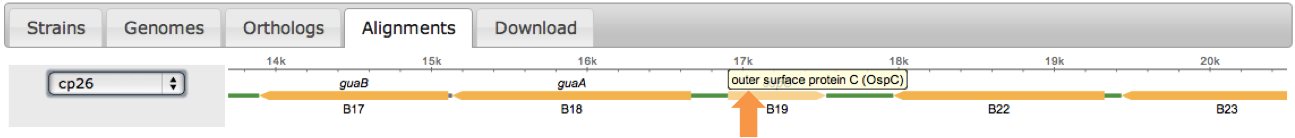
Once selected, a scrollable map will appear. Use your browser default controls to search (ctrl/cmd + f) to find specific genes or scroll using the mouse. Select the gene of interest by clicking on the orange rectangle, hovering over these features will reveal annotations.
- Orange -> Open Reading Frame (ORF)
- Green -> Inter Genetic Sequence (IGS)
- Gray -> No Information
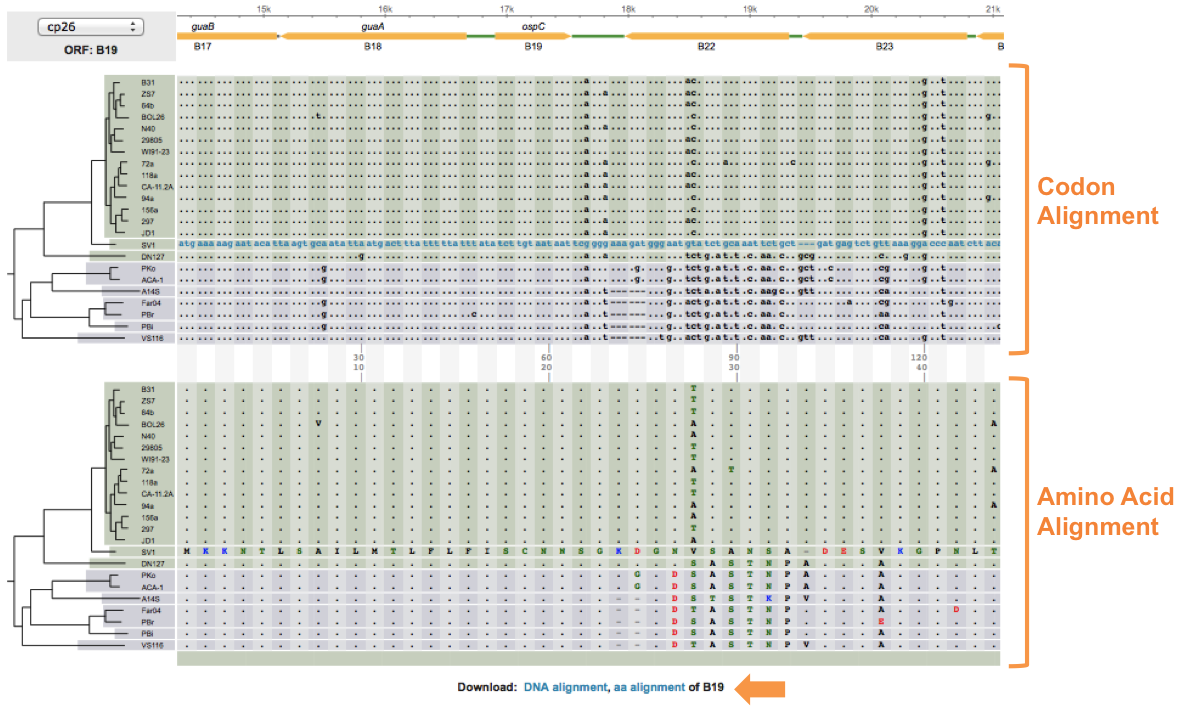
Select a gene or intergenic region by clicking on the corresponding rectangle (Green for IGS, Orange for orthologous ORF). Two phylogenetic trees will become populated with the alignment for the region selected. The top tree shows the codon alignment where as the bottom shows the corresponding amino acid alignment. These maps are generated on the fly using MUSCLE. Additionally, these alignments can be downloaded in CLUSTALW format by clicking on the links on the bottom of the page.