Pseudomonas population genomics: Difference between revisions
Jump to navigation
Jump to search
imported>Rayrah |
imported>Rayrah |
||
| Line 18: | Line 18: | ||
## Protocol: | ## Protocol: | ||
### Fasta headers are too long for tree, run: [https://www.dropbox.com/s/x2p4joeqg7omfub/rename.pl rename.pl] to shorten names. Usage:<code> >rename.pl <FASTA_file> > <OUTPUTfilename.fas> </code> (will rewrite script to create automatic output file) | ### Fasta headers are too long for tree, run: [https://www.dropbox.com/s/x2p4joeqg7omfub/rename.pl rename.pl] to shorten names. Usage:<code> >rename.pl <FASTA_file> > <OUTPUTfilename.fas> </code> (will rewrite script to create automatic output file) | ||
### To align use [http://www.drive5.com/muscle/Alignment muscle] Usage: <code> >muscle -in <FASTA_File> -out <OUTPUTfilename | ### To align use [http://www.drive5.com/muscle/Alignment muscle] Usage: <code> >muscle -in <FASTA_File> -out <OUTPUTfilename> -clwstrict </code> | ||
### To create tree file run [http://www.clustal.org/clustal2/ clustalW2] Usage <code>>clustalw2 -infile= <Aligned_file.aln> -output= Phylip </code> | ### To create tree file run [http://www.clustal.org/clustal2/ clustalW2] Usage <code>>clustalw2 -infile= <Aligned_file.aln> -output= Phylip </code> | ||
### To create tree run [http://www.r-project.org/ R] using following commands: | ### To create tree run [http://www.r-project.org/ R] using following commands: | ||
Revision as of 21:21, 18 June 2013
Projects
- Build a local genome database
- Database schema:
- "genome": genome_id, strain_name, ncbi_taxid
- "orf": genome_id, locus_tag, start, stop, strand, genome_name, product_name
- "orth_orf": orth_orf_id, locus_name, genome_id, orth_class
- Parsing scripts
- Rayees Parsing code, requires that you remove columns 9-27 using bash command:
cut -c 1-8(I will write a bash script that does this and runs the program) https://www.dropbox.com/s/lpxxbkxeyw7frrn/parser.pl
- Rayees Parsing code, requires that you remove columns 9-27 using bash command:
- Database loading scripts
- Database schema:
- Molecular Evolution of flagellum genes
- Download orthologs
- Reconstruct phylogenetic tree
- Run PAML tests
Update: June 18, 2013
- Material and Methods
- Protein ortholog data: fleN, fleQ, flhF
- Protocol:
- Fasta headers are too long for tree, run: rename.pl to shorten names. Usage:
>rename.pl <FASTA_file> > <OUTPUTfilename.fas>(will rewrite script to create automatic output file) - To align use muscle Usage:
>muscle -in <FASTA_File> -out <OUTPUTfilename> -clwstrict - To create tree file run clustalW2 Usage
>clustalw2 -infile= <Aligned_file.aln> -output= Phylip - To create tree run R using following commands:
setwd("<directory containing phylip files>")library ("ape")library ("phangorn")<gene_name> = read.tree("<file_gene_name.phy>")<gene_name> = midpoint(gene_name)plot(<gene_name>)
- Fasta headers are too long for tree, run: rename.pl to shorten names. Usage:
| Gene | Alignment | Tree | Notes |
|---|---|---|---|
| fleN | fleN pep alignment | 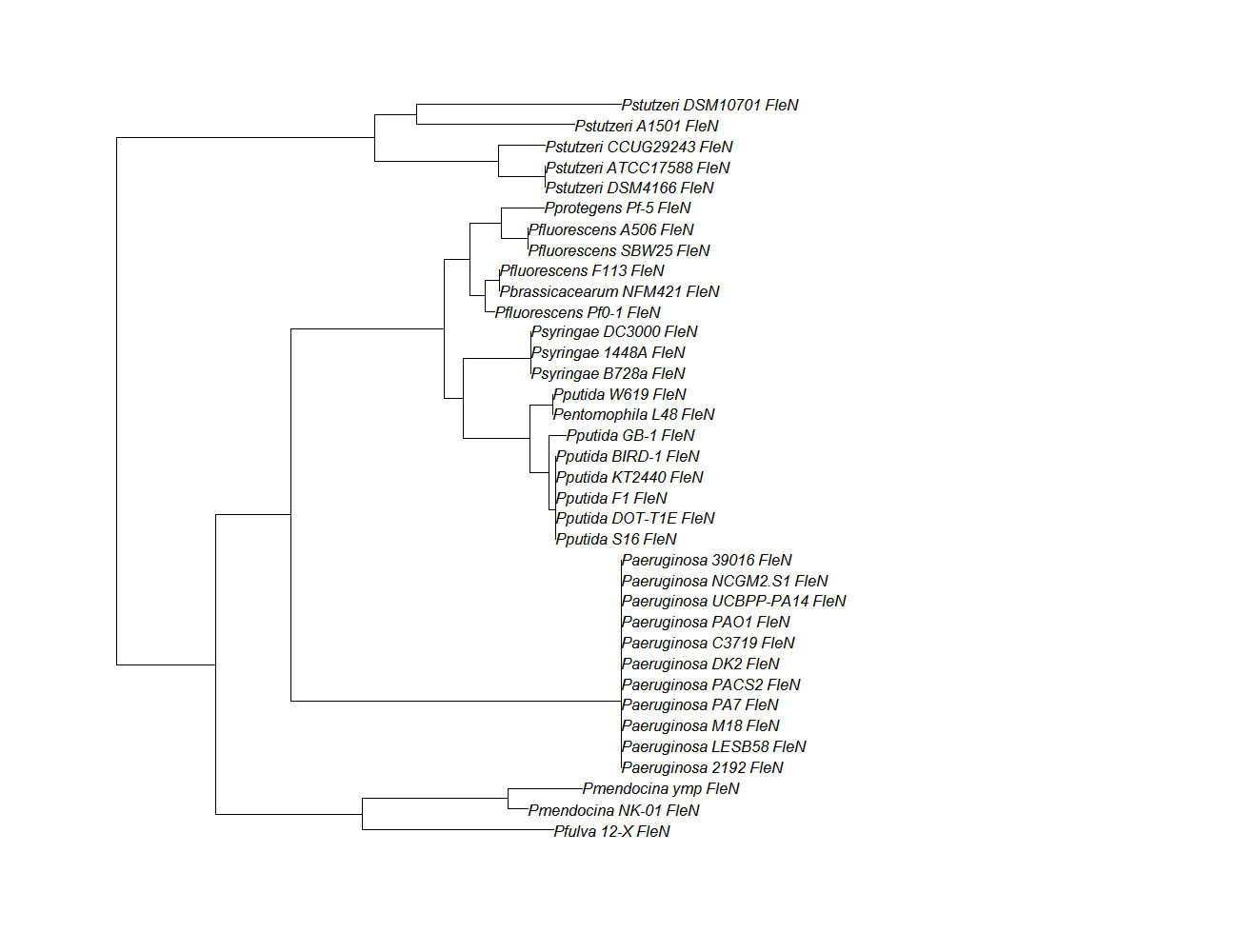 |
Conserved Domain |
| fleQ | fleQ pep alignment | 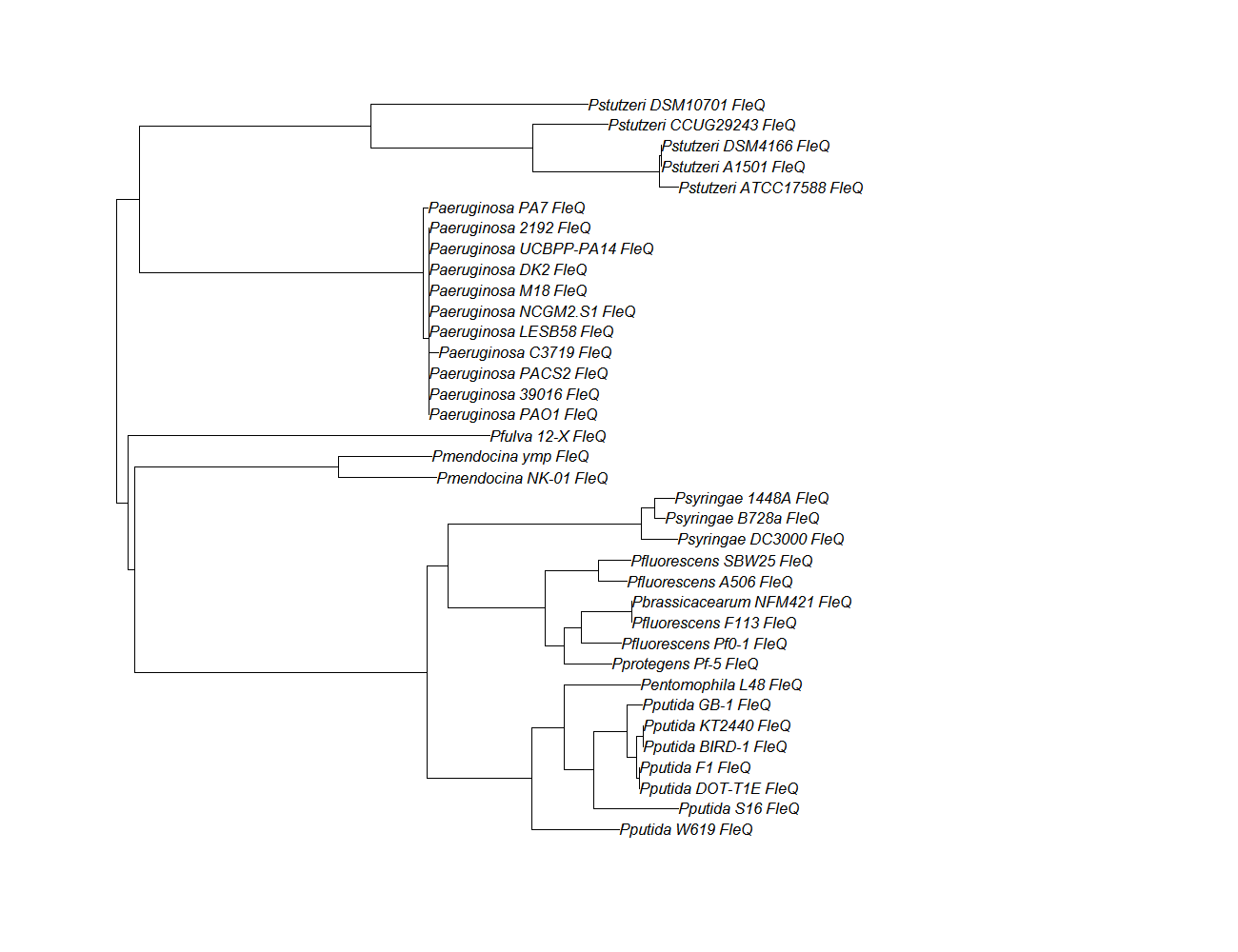 |
Conserved Domain |
| flhf | flhF pep alignment | 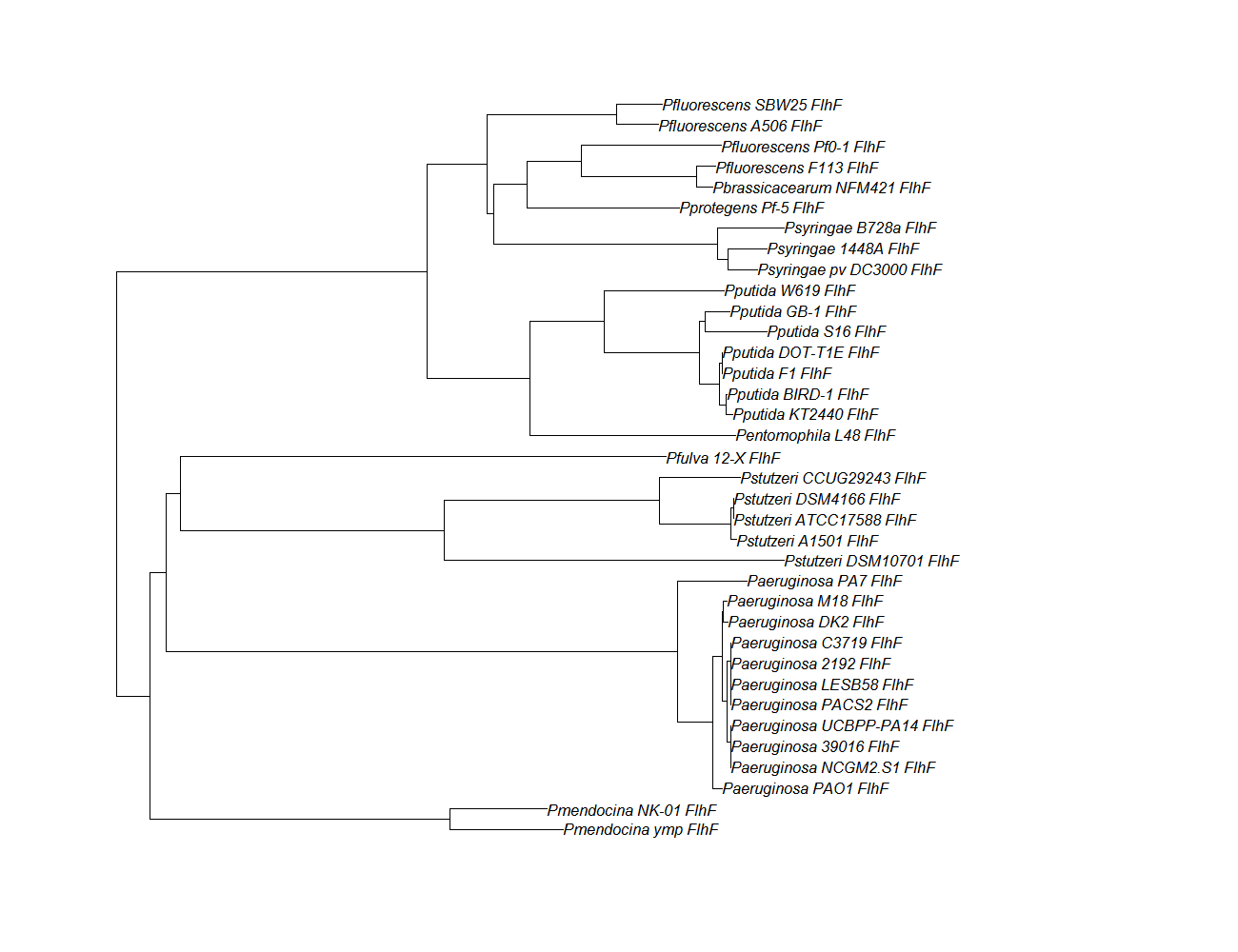 |
Conserved Domain |
Benchmark: June 11, 2013
- Finish parsing the genome files to upload the "orf" table (Raymond & Rayees)
- Rayees Parsed genome files: https://www.dropbox.com/sh/k0zktvvmv39op9i/1zBercEky8
- Parsing the ortholog file to upload the "orth_orf" table (Raymond)
- Identify and download fleN, fleQ, and flhF orthologs & align them (Rayees)