QuBi/modules/biol302: Difference between revisions
imported>Weigang |
imported>Weigang mNo edit summary |
||
| Line 8: | Line 8: | ||
*[[TODO: Link to BIOL302 page, if one exists]] | *[[TODO: Link to BIOL302 page, if one exists]] | ||
==[[QuBi/modules/biol302#Part 1: Identification of mdm2 Splice Variants Using BLAST|Part 1: Identification of mdm2 Splice Variants Using BLAST]]== | |||
==[[QuBi/modules/biol302#MODULE: Cloning of murine mdm2 gene sequence to study cis acting DNA elements|Part 1: Cloning of murine mdm2 gene sequence to study cis acting DNA elements]]== | |||
[[TODO: Summarize project ]] | |||
===[[QuBi/modules/biol302#Key Concepts2|Key Concepts]]=== | |||
*Homology searching using BLAST | |||
*Mammalian gene enhancers | |||
===[[QuBi/modules/biol302#Exercise2|Exercise]]=== | |||
So far, you have excised this XbaI fragment: | |||
<div style="font-family:Monospace;line-height:1;width:550px;border-style:solid;border-width:1px;border-color:#AAAAFF;background-color:#EEEEFF;padding-left:5px;padding-right:5px;padding-top:0px;padding-bottom:0px;"> | |||
TCTAGATGCATTTACGAAGGAGACAGAAAACGTCTTTCGGCAATAGCTCTCAAATGCAAAACGACGTCGG | |||
CGAGCTGTCCCTTACCTGGAGGCCCGCAGGAGAAGCGCGGTGATCCGAGAGGGTCCCCCAGGGGTGTCCG | |||
GTCGGTCTCCCGCTCGCCCAGCAGACGGCTGCGGAAACGGGGCAGCGTTTAAATAACCCCAGCTGGAGAC | |||
ATGTCAGGACTTAGCTCCTCCGACAGCCGACGCCGGACGTGTCCCAACTTGACCAGCCCCACAGGAAGAG | |||
CTGAGTCAACTCGGCCCAGCCCAGTCCCACCCGTCCCGGAAGCCGCATCCCGGCGAGTCCGGGACCAGGC | |||
ACCTGTCACCTCCTGGACCCCAGCAACGAGCCCAGCGCGACCCCGGAGCGGGCCCGAATTCTCTAGA | |||
</div> | |||
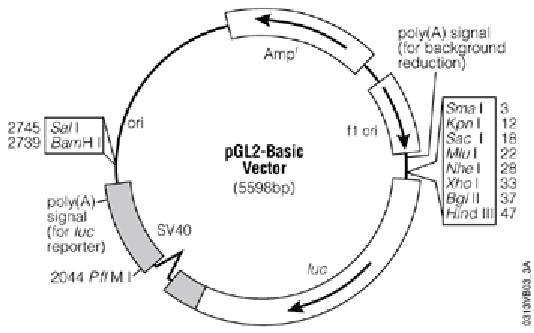
and cloned it into the NheI site in the pGL2basic vector [[vector]]: | |||
<div style="font-family:Monospace;line-height:1;width:400px;border-style:solid;border-width:1px;border-color:#AAAAFF;background-color:#EEEEFF;padding-left:5px;padding-right:5px;padding-top:0px;padding-bottom:10px;"> | |||
[[File:PGL2_basic.png|400px]] | |||
''[http://www.promega.com/products/reporter-assays-and-transfection/reporter-vectors-and-cell-lines/pgl2-luciferase-reporter-vectors/ pGL2 Luciferase Reporter Vectors]'' | |||
</div> | |||
Exercise 1. Identify the gene in the mouse genome that is associate with this sequence. | |||
* Software Tool: BLAST | |||
* Protocol | |||
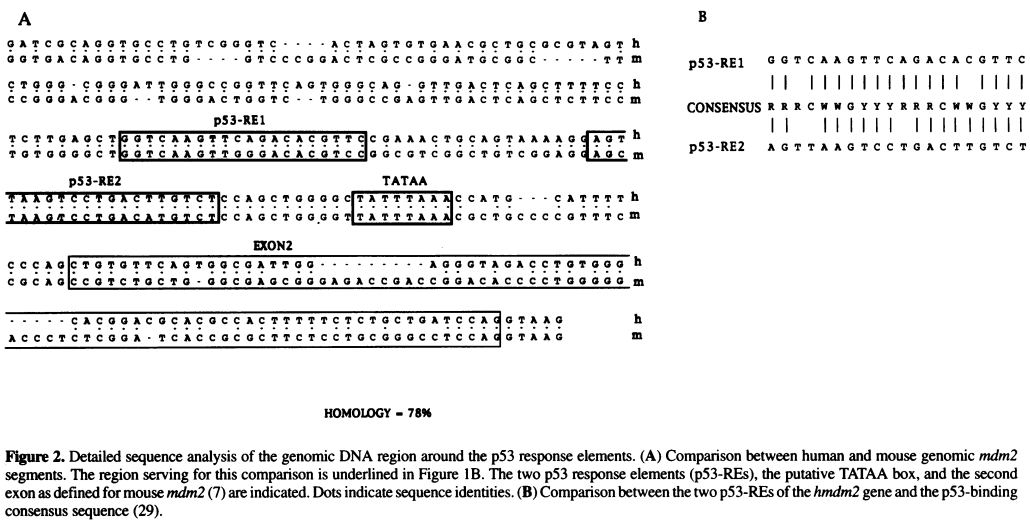
Exercise 2. Identify the TATA box, p52 RE1, p52 RE2, and exon 2 sequences by comparing with this diagram: | |||
<div style="font-family:Monospace;line-height:1;width:800px;border-style:solid;border-width:1px;border-color:#AAAAFF;background-color:#EEEEFF;padding-left:5px;padding-right:5px;padding-top:0px;padding-bottom:0px;"> | |||
[[File:Zauberman_fig2.png|800px]] | |||
[[PROPERLY CITE THIS]] ''Taken from '' A functional p53-responsive intronic promoter is contained within the human mdm2 gene. ''[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC307078/ Pubmed] [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC307078/pdf/nar00014-0022.pdf PDF]'' | |||
</div> | |||
---- | |||
; Annotate fragment and draw diagram of resultant clone | |||
This exercise is not bioinformatics-heavy, but sequence analysis is a major task of the field. You will create another annotated drawing of a DNA molecule (the plasmid with your fragment cloned in) and annotate your fragment sequence by hand. You should have a printout of the fragment sequence to directly annotate. | |||
# On your sequence, | |||
[[BRAINSTORMING]] | |||
The students could blast their fragment and give exactly where the sequence is from, provide the accession number (either JN963162.1 or JN955500.1), list the range of bases that match, and explain what the non-matching side bases are (XbaI/EcoRI sites).They can view the graphics of the alignment. Hovering over the "BLAST Results for:" track, it will show that the first 5 and last 12 bases do not align. They can go back to their original sequence and annotate each section with the accession number, base-match range, and restriction sites. | |||
[[/BRAINSTORMING]] | |||
==[[QuBi/modules/biol302#Part 2: Identification of mdm2 Splice Variants Using BLAST|Part 1: Identification of mdm2 Splice Variants Using BLAST]]== | |||
<div class="center"> | <div class="center"> | ||
[[File:4seq.png|800px]] | [[File:4seq.png|800px]] | ||
| Line 15: | Line 64: | ||
</div> | </div> | ||
---- | ---- | ||
===[[QuBi/modules/biol302#Objectives|Objectives]]=== | ===[[QuBi/modules/biol302#Objectives|Objectives]]=== | ||
* Learn to use Genbank database and BLAST tool to analyze nucleotide sequences | * Learn to use Genbank database and BLAST tool to analyze nucleotide sequences | ||
| Line 70: | Line 122: | ||
<!-- ===[[QuBi/modules/biol302#Exit_Questions|Exit Questions]]=== --> | <!-- ===[[QuBi/modules/biol302#Exit_Questions|Exit Questions]]=== --> | ||
Revision as of 22:06, 18 March 2013
- BIOL 302 Lab (Bioinformatics Exercises)
Research in molecular genetics requires effective use of bioinformatic tools to analyze and understand the genetic materials being worked with. The following exercises will expose you to real-world scenarios and introduce you to the methods and tools you can use to solve these problems.
Part 1: Cloning of murine mdm2 gene sequence to study cis acting DNA elements
Key Concepts
- Homology searching using BLAST
- Mammalian gene enhancers
Exercise
So far, you have excised this XbaI fragment:
TCTAGATGCATTTACGAAGGAGACAGAAAACGTCTTTCGGCAATAGCTCTCAAATGCAAAACGACGTCGG CGAGCTGTCCCTTACCTGGAGGCCCGCAGGAGAAGCGCGGTGATCCGAGAGGGTCCCCCAGGGGTGTCCG GTCGGTCTCCCGCTCGCCCAGCAGACGGCTGCGGAAACGGGGCAGCGTTTAAATAACCCCAGCTGGAGAC ATGTCAGGACTTAGCTCCTCCGACAGCCGACGCCGGACGTGTCCCAACTTGACCAGCCCCACAGGAAGAG CTGAGTCAACTCGGCCCAGCCCAGTCCCACCCGTCCCGGAAGCCGCATCCCGGCGAGTCCGGGACCAGGC ACCTGTCACCTCCTGGACCCCAGCAACGAGCCCAGCGCGACCCCGGAGCGGGCCCGAATTCTCTAGA
and cloned it into the NheI site in the pGL2basic vector vector:
Exercise 1. Identify the gene in the mouse genome that is associate with this sequence.
- Software Tool: BLAST
- Protocol
Exercise 2. Identify the TATA box, p52 RE1, p52 RE2, and exon 2 sequences by comparing with this diagram:
PROPERLY CITE THIS Taken from A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Pubmed PDF
- Annotate fragment and draw diagram of resultant clone
This exercise is not bioinformatics-heavy, but sequence analysis is a major task of the field. You will create another annotated drawing of a DNA molecule (the plasmid with your fragment cloned in) and annotate your fragment sequence by hand. You should have a printout of the fragment sequence to directly annotate.
- On your sequence,
The students could blast their fragment and give exactly where the sequence is from, provide the accession number (either JN963162.1 or JN955500.1), list the range of bases that match, and explain what the non-matching side bases are (XbaI/EcoRI sites).They can view the graphics of the alignment. Hovering over the "BLAST Results for:" track, it will show that the first 5 and last 12 bases do not align. They can go back to their original sequence and annotate each section with the accession number, base-match range, and restriction sites.
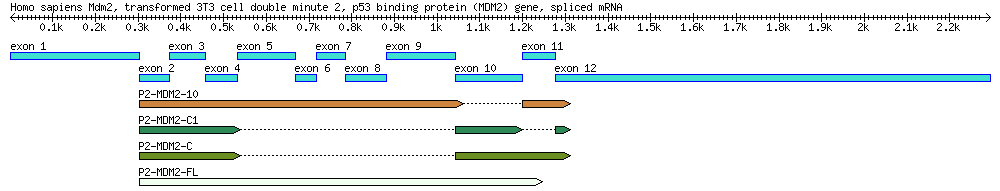
Part 1: Identification of mdm2 Splice Variants Using BLAST
A diagram of the MDM2 gene used in this exercise, along with its splice variants. By the end of this module you will create a similar diagram.
Objectives
- Learn to use Genbank database and BLAST tool to analyze nucleotide sequences
- Use BLAST to identify
Key Concepts
- Blast
- Genbank
- Annotation
- Accession Number
- Alternative Splicing
Exercise
| Genbank Accession # | cDNA Clone | Description | Cell Line | Length (bp) |
|---|---|---|---|---|
| AF527840 | Genomic DNA | 34,088 | ||
| EU076746 | P2-MDM2-C1 | cDNA missing exons 5-9 & 11 | MANCA | 427 |
| EU076747 | P2-MDM2-10 | cDNA missing exon 10 | ML-1 | 842 |
| EU076748 | P2-MDM2-C | cDNA missing exons 5-9 | A876 | 505 |
| EU076749 | P2-MDM2-FL | Full-length cDNA | SJSA-1 | 845 |
- Explore the gene annotation for AF527840.
Sequences on genbank have both basic reference information (such as what the sequence is, what organism it came from, and bibliographical information) and sequence annotations. Some sequences are more richly annotated than others - it is up to researchers to annotate the sequences they generate, which requires extra work. For this exercise you will be working will a well-annotated sequence: accession number AF527840. Explore its annotation and use it to complete the following set of tasks.
- DRAW a diagram of this gene using the information and coordinates listed in the annotation. (Note: this is the bulk of the assignment and this diagram is needed for the last set of questions. Don't get lazy on this.)
- Label the top of the diagram with basic information, such as the gene's name, organism, etc.. Someone should be able to pick up your diagram and know exactly what they're looking at.
- Including introns, exons, 3'/5' UTRs, +1, and exact coordinates. (The mRNA annotation states which segments are used to create mRNA, and the CDS annotation states which parts code amino acids (CDS = coding sequence)).
- Draw the diagram mostly to scale. It does NOT have to be perfect, but make a reasonable effort. Put a scale bar and length markers on your drawing.
- How does the sequence vary at positions X, X, and X for this gene? Do these change the AA for the resulting peptide?
- What kinds of repeat regions can be found in this gene?
- Explore the graphical presentation of the gene to answer the following question.
Genbank provides graphical representations of the sequences on its database: click the "Graphics" link below the sequence title, OR click "Display Settings" above the title, and choose "Graphics". Take a few minutes to explore this graphical browser and answer the following question:
- A question that can only be answered from looking at this graph.
- Another question that can only be answered from looking at this graph.
- Use BLAST to determine which exons are used in the mRNA transcripts.
This is the most "bioinformatic" part of the assignment. Blast ALL FOUR of the mRNA sequences (EU076746, EU076747, EU076748, EU076749) against the main sequence (AF527840) and use the results to answer the following questions. If you labeled your diagram well (with coordinates!), this task should go by quickly.
- Which exons are used to create EU076746, EU076747, EU076748, EU076749?
- Do the BLAST search results corresponding to exons exactly match the start/end positions of the exons as labeled in your diagram? If not, what is the most likely reason for this?
- Do any of the BLAST results match regions outside of exons? If so, what regions?