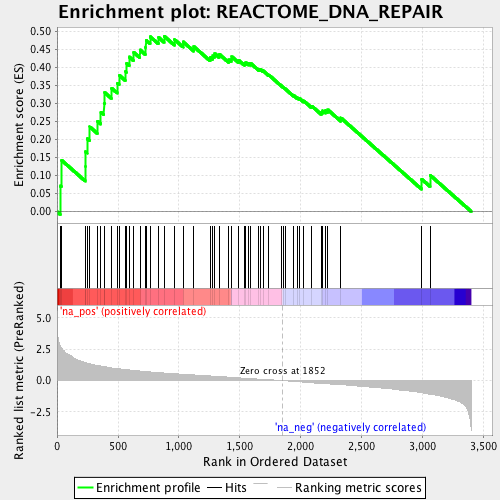
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | chrom |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_DNA_REPAIR |
| Enrichment Score (ES) | 0.48722365 |
| Normalized Enrichment Score (NES) | 1.8130012 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.022902012 |
| FWER p-Value | 0.173 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MPG | MPG Entrez, Source | N-methylpurine-DNA glycosylase | 27 | 2.762 | 0.0713 | Yes |
| 2 | PCNA | PCNA Entrez, Source | proliferating cell nuclear antigen | 36 | 2.572 | 0.1428 | Yes |
| 3 | ERCC5 | ERCC5 Entrez, Source | me)) | 231 | 1.428 | 0.1259 | Yes |
| 4 | RPA2 | RPA2 Entrez, Source | replication protein A2, 32kDa | 234 | 1.421 | 0.1661 | Yes |
| 5 | MNAT1 | MNAT1 Entrez, Source | menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 247 | 1.393 | 0.2025 | Yes |
| 6 | POLR2G | POLR2G Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide G | 268 | 1.353 | 0.2354 | Yes |
| 7 | MBD4 | MBD4 Entrez, Source | methyl-CpG binding domain protein 4 | 331 | 1.197 | 0.2513 | Yes |
| 8 | TDP1 | TDP1 Entrez, Source | tyrosyl-DNA phosphodiesterase 1 | 360 | 1.145 | 0.2758 | Yes |
| 9 | NTHL1 | NTHL1 Entrez, Source | nth endonuclease III-like 1 (E. coli) | 385 | 1.105 | 0.3004 | Yes |
| 10 | POLR2I | POLR2I Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa | 391 | 1.097 | 0.3305 | Yes |
| 11 | RFC4 | RFC4 Entrez, Source | replication factor C (activator 1) 4, 37kDa | 447 | 1.003 | 0.3429 | Yes |
| 12 | ATR | ATR Entrez, Source | ataxia telangiectasia and Rad3 related | 494 | 0.950 | 0.3564 | Yes |
| 13 | RFC2 | RFC2 Entrez, Source | replication factor C (activator 1) 2, 40kDa | 511 | 0.930 | 0.3784 | Yes |
| 14 | MDC1 | MDC1 Entrez, Source | mediator of DNA damage checkpoint 1 | 561 | 0.882 | 0.3891 | Yes |
| 15 | MAD2L2 | MAD2L2 Entrez, Source | MAD2 mitotic arrest deficient-like 2 (yeast) | 570 | 0.870 | 0.4117 | Yes |
| 16 | RFC3 | RFC3 Entrez, Source | replication factor C (activator 1) 3, 38kDa | 594 | 0.851 | 0.4293 | Yes |
| 17 | POLR2C | POLR2C Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide C, 33kDa | 629 | 0.809 | 0.4423 | Yes |
| 18 | POLR2F | POLR2F Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide F | 681 | 0.759 | 0.4489 | Yes |
| 19 | XRCC1 | XRCC1 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 1 | 725 | 0.716 | 0.4566 | Yes |
| 20 | XAB2 | XAB2 Entrez, Source | XPA binding protein 2 | 730 | 0.713 | 0.4759 | Yes |
| 21 | RFC5 | RFC5 Entrez, Source | replication factor C (activator 1) 5, 36.5kDa | 764 | 0.685 | 0.4857 | Yes |
| 22 | DDB2 | DDB2 Entrez, Source | damage-specific DNA binding protein 2, 48kDa | 833 | 0.630 | 0.4835 | Yes |
| 23 | CDK7 | CDK7 Entrez, Source | cyclin-dependent kinase 7 (MO15 homolog, Xenopus laevis, cdk-activating kinase) | 879 | 0.597 | 0.4872 | Yes |
| 24 | DDB1 | DDB1 Entrez, Source | damage-specific DNA binding protein 1, 127kDa | 964 | 0.543 | 0.4777 | No |
| 25 | POLR2D | POLR2D Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide D | 1035 | 0.507 | 0.4714 | No |
| 26 | LIG3 | LIG3 Entrez, Source | ligase III, DNA, ATP-dependent | 1123 | 0.447 | 0.4582 | No |
| 27 | POLR2A | POLR2A Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | 1258 | 0.369 | 0.4288 | No |
| 28 | RPA1 | RPA1 Entrez, Source | replication protein A1, 70kDa | 1280 | 0.355 | 0.4327 | No |
| 29 | MRE11A | MRE11A Entrez, Source | MRE11 meiotic recombination 11 homolog A (S. cerevisiae) | 1295 | 0.347 | 0.4385 | No |
| 30 | POLR2B | POLR2B Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide B, 140kDa | 1331 | 0.322 | 0.4373 | No |
| 31 | POLR2E | POLR2E Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | 1411 | 0.275 | 0.4216 | No |
| 32 | XRCC6 | XRCC6 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa) | 1431 | 0.264 | 0.4235 | No |
| 33 | POLD2 | POLD2 Entrez, Source | polymerase (DNA directed), delta 2, regulatory subunit 50kDa | 1435 | 0.261 | 0.4301 | No |
| 34 | POLB | POLB Entrez, Source | polymerase (DNA directed), beta | 1492 | 0.223 | 0.4198 | No |
| 35 | CCNH | CCNH Entrez, Source | cyclin H | 1539 | 0.187 | 0.4114 | No |
| 36 | POLR2H | POLR2H Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide H | 1550 | 0.183 | 0.4136 | No |
| 37 | PRKDC | PRKDC Entrez, Source | protein kinase, DNA-activated, catalytic polypeptide | 1574 | 0.167 | 0.4116 | No |
| 38 | FEN1 | FEN1 Entrez, Source | flap structure-specific endonuclease 1 | 1592 | 0.158 | 0.4110 | No |
| 39 | XRCC5 | XRCC5 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | 1658 | 0.115 | 0.3949 | No |
| 40 | POLR2L | POLR2L Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa | 1670 | 0.109 | 0.3947 | No |
| 41 | POLR2K | POLR2K Entrez, Source | polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa | 1692 | 0.094 | 0.3912 | No |
| 42 | RPS27A | RPS27A Entrez, Source | ribosomal protein S27a | 1739 | 0.066 | 0.3793 | No |
| 43 | MGMT | MGMT Entrez, Source | O-6-methylguanine-DNA methyltransferase | 1843 | 0.007 | 0.3488 | No |
| 44 | XRCC4 | XRCC4 Entrez, Source | X-ray repair complementing defective repair in Chinese hamster cells 4 | 1859 | -0.004 | 0.3444 | No |
| 45 | TP53BP1 | TP53BP1 Entrez, Source | tumor protein p53 binding protein, 1 | 1872 | -0.014 | 0.3412 | No |
| 46 | ERCC3 | ERCC3 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 3 (xeroderma pigmentosum group B complementing) | 1944 | -0.069 | 0.3220 | No |
| 47 | POLD1 | POLD1 Entrez, Source | polymerase (DNA directed), delta 1, catalytic subunit 125kDa | 1974 | -0.087 | 0.3158 | No |
| 48 | RAD50 | RAD50 Entrez, Source | RAD50 homolog (S. cerevisiae) | 1991 | -0.099 | 0.3139 | No |
| 49 | GTF2H4 | GTF2H4 Entrez, Source | general transcription factor IIH, polypeptide 4, 52kDa | 2025 | -0.126 | 0.3076 | No |
| 50 | LIG4 | LIG4 Entrez, Source | ligase IV, DNA, ATP-dependent | 2092 | -0.175 | 0.2929 | No |
| 51 | NBN | NBN Entrez, Source | nibrin | 2171 | -0.226 | 0.2761 | No |
| 52 | RPA3 | RPA3 Entrez, Source | replication protein A3, 14kDa | 2182 | -0.233 | 0.2798 | No |
| 53 | GTF2H3 | GTF2H3 Entrez, Source | general transcription factor IIH, polypeptide 3, 34kDa | 2204 | -0.248 | 0.2807 | No |
| 54 | ERCC2 | ERCC2 Entrez, Source | excision repair cross-complementing rodent repair deficiency, complementation group 2 (xeroderma pigmentosum D) | 2224 | -0.262 | 0.2825 | No |
| 55 | APEX1 | APEX1 Entrez, Source | APEX nuclease (multifunctional DNA repair enzyme) 1 | 2331 | -0.324 | 0.2601 | No |
| 56 | GTF2H1 | GTF2H1 Entrez, Source | general transcription factor IIH, polypeptide 1, 62kDa | 2994 | -0.969 | 0.0901 | No |
| 57 | TCEA1 | TCEA1 Entrez, Source | transcription elongation factor A (SII), 1 | 3064 | -1.097 | 0.1010 | No |