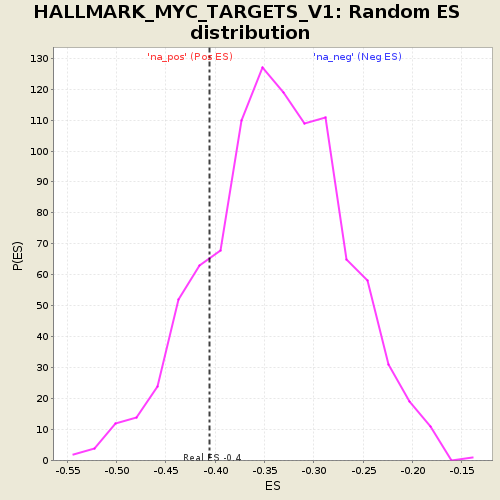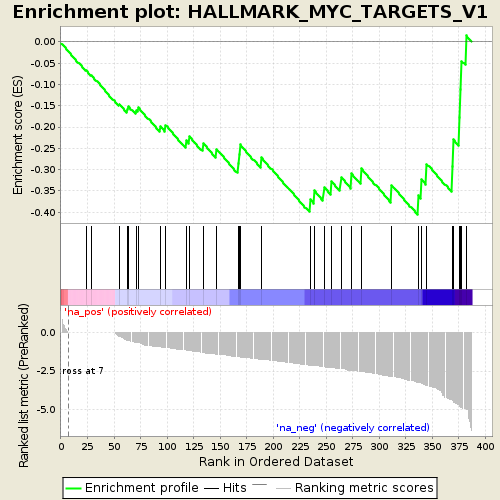
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | list_small_change_phospho_genes |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | HALLMARK_MYC_TARGETS_V1 |
| Enrichment Score (ES) | -0.4057043 |
| Normalized Enrichment Score (NES) | -1.2006011 |
| Nominal p-value | 0.17 |
| FDR q-value | 0.54349893 |
| FWER p-Value | 0.445 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EPRS | EPRS Entrez, Source | glutamyl-prolyl-tRNA synthetase | 24 | 0.000 | -0.0678 | No |
| 2 | CBX3 | CBX3 Entrez, Source | chromobox homolog 3 (HP1 gamma homolog, Drosophila) | 29 | 0.000 | -0.0791 | No |
| 3 | SRSF2 | 55 | -0.258 | -0.1462 | No | ||
| 4 | SRSF7 | 63 | -0.501 | -0.1591 | No | ||
| 5 | SRSF3 | 64 | -0.512 | -0.1522 | No | ||
| 6 | SRSF1 | 71 | -0.644 | -0.1603 | No | ||
| 7 | DEK | DEK Entrez, Source | DEK oncogene (DNA binding) | 73 | -0.646 | -0.1544 | No |
| 8 | RRP9 | 94 | -0.914 | -0.1984 | No | ||
| 9 | HNRNPD | 99 | -0.958 | -0.1967 | No | ||
| 10 | NPM1 | NPM1 Entrez, Source | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 118 | -1.130 | -0.2321 | No |
| 11 | RPL14 | RPL14 Entrez, Source | ribosomal protein L14 | 121 | -1.154 | -0.2221 | No |
| 12 | TRA2B | 134 | -1.290 | -0.2384 | No | ||
| 13 | HNRNPA1 | 147 | -1.398 | -0.2532 | No | ||
| 14 | HNRNPU | 167 | -1.581 | -0.2854 | No | ||
| 15 | SSB | SSB Entrez, Source | Sjogren syndrome antigen B (autoantigen La) | 168 | -1.584 | -0.2638 | No |
| 16 | HNRNPC | 169 | -1.592 | -0.2421 | No | ||
| 17 | EIF2S2 | EIF2S2 Entrez, Source | eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa | 189 | -1.726 | -0.2723 | No |
| 18 | HNRNPA3 | 235 | -2.133 | -0.3703 | Yes | ||
| 19 | PWP1 | PWP1 Entrez, Source | PWP1 homolog (S. cerevisiae) | 239 | -2.156 | -0.3494 | Yes |
| 20 | HDGF | HDGF Entrez, Source | hepatoma-derived growth factor (high-mobility group protein 1-like) | 248 | -2.233 | -0.3416 | Yes |
| 21 | RPS3 | RPS3 Entrez, Source | ribosomal protein S3 | 255 | -2.285 | -0.3275 | Yes |
| 22 | RAD23B | RAD23B Entrez, Source | RAD23 homolog B (S. cerevisiae) | 264 | -2.349 | -0.3181 | Yes |
| 23 | PRPF31 | PRPF31 Entrez, Source | PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae) | 274 | -2.475 | -0.3098 | Yes |
| 24 | RSL1D1 | RSL1D1 Entrez, Source | ribosomal L1 domain containing 1 | 283 | -2.535 | -0.2978 | Yes |
| 25 | CANX | CANX Entrez, Source | calnexin | 312 | -2.866 | -0.3379 | Yes |
| 26 | HDAC2 | HDAC2 Entrez, Source | histone deacetylase 2 | 337 | -3.233 | -0.3617 | Yes |
| 27 | MCM2 | MCM2 Entrez, Source | MCM2 minichromosome maintenance deficient 2, mitotin (S. cerevisiae) | 340 | -3.282 | -0.3226 | Yes |
| 28 | HNRNPA2B1 | 345 | -3.437 | -0.2871 | Yes | ||
| 29 | NOLC1 | NOLC1 Entrez, Source | nucleolar and coiled-body phosphoprotein 1 | 369 | -4.418 | -0.2919 | Yes |
| 30 | TRIM28 | TRIM28 Entrez, Source | tripartite motif-containing 28 | 370 | -4.555 | -0.2299 | Yes |
| 31 | PCBP1 | PCBP1 Entrez, Source | poly(rC) binding protein 1 | 376 | -4.786 | -0.1788 | Yes |
| 32 | DDX21 | DDX21 Entrez, Source | DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | 377 | -4.866 | -0.1125 | Yes |
| 33 | TOMM70A | TOMM70A Entrez, Source | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | 378 | -4.901 | -0.0458 | Yes |
| 34 | NOP56 | 382 | -5.018 | 0.0141 | Yes |