Main Page: Difference between revisions
Jump to navigation
Jump to search
| (36 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
|- | |- | ||
| | | | ||
[[File: | [[File:Protrait-July-2024-Qiu.jpg| x250 px | thumb]] | ||
| <center> <strong>Welcome to Qiu Lab Wiki @ Hunter</strong><br>Weigang Qiu, Ph.D., Professor | | <center> <strong>Welcome to Qiu Lab Wiki @ Hunter</strong><br>Weigang Qiu, Ph.D., Professor, [http://biology.hunter.cuny.edu Department of Biological Sciences]<br> | ||
[https://hunter.cuny.edu Hunter College] of | [https://hunter.cuny.edu Hunter College] of [https://www.cuny.edu City University of New York]<br> | ||
Adjunct Faculty, Weill Cornell Medical College, Department of Physiology and Biophysics<br> | |||
Belfer Research Building, Room 402<br>413 East 69th Street, New York, NY 10021<br>Office: 1-212-896-0445<br>Email: wqiu-at-(hunter.cuny.edu)<br> </center> | |||
| [[File:Belfer_building.jpg | x300px | thumb | [https://goo.gl/maps/xv1KmaW3XEnxaY1V7 Directions by Google Map]]] | | [[File:Belfer_building.jpg | x300px | thumb | [https://goo.gl/maps/xv1KmaW3XEnxaY1V7 Directions by Google Map]]] | ||
|__TOC__ | |__TOC__ | ||
| Line 20: | Line 22: | ||
===Lyme Genomics, Evolution, & Ecology=== | ===Lyme Genomics, Evolution, & Ecology=== | ||
<gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | <gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | ||
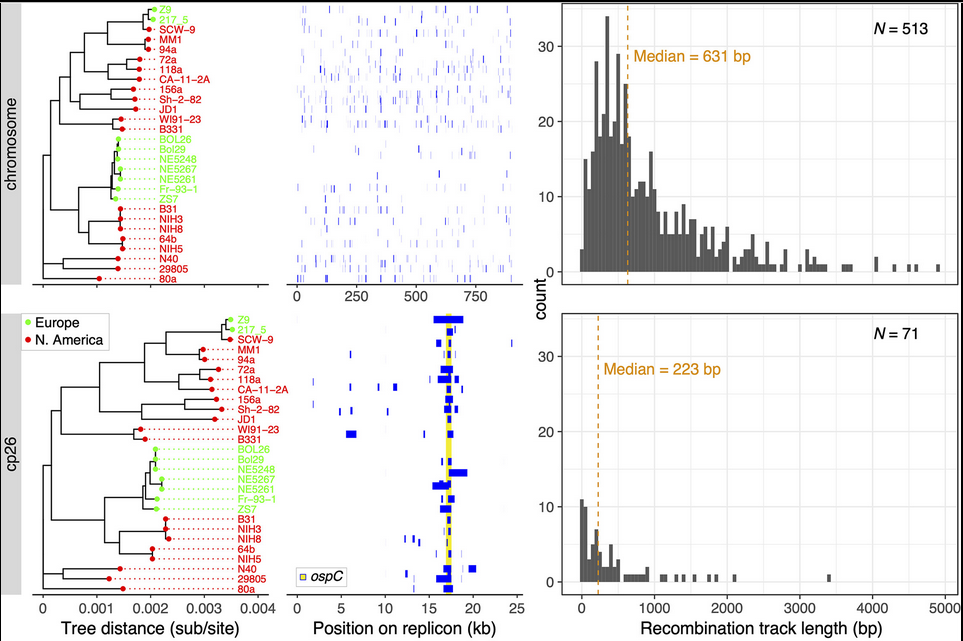
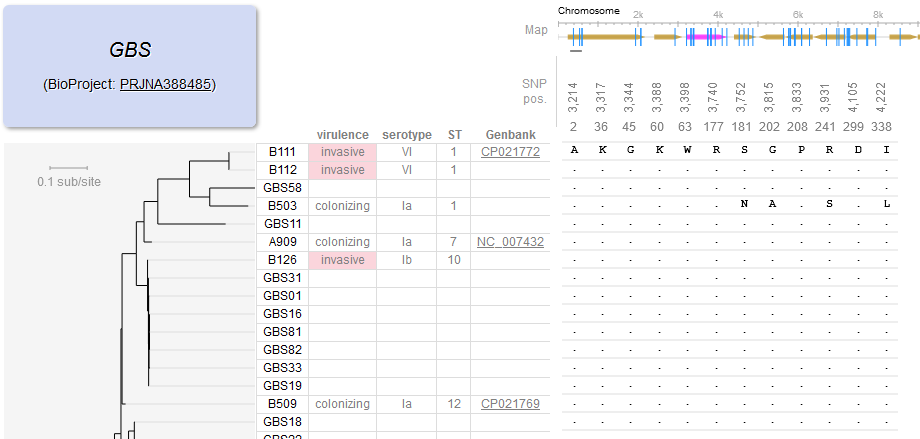
File:Screenshot_2024-08-15_094908.png | link=https://journals.asm.org/doi/10.1128/mbio.01749-24 | Akther et al. 2024. "Natural selection and recombination at host-interacting lipoprotein loci drive genome diversification of Lyme disease and related bacteria" '''''mBio''''' 0:e01749-24. Press coverage: [https://www.gc.cuny.edu/news/cuny-graduate-center-biologists-map-dna-lyme-disease-bacteria CUNY Graduate Center News Story]; [https://hunter.cuny.edu/news/hunter-researcher-maps-dna-of-lyme-disease-bacteria/ Hunter press] | |||
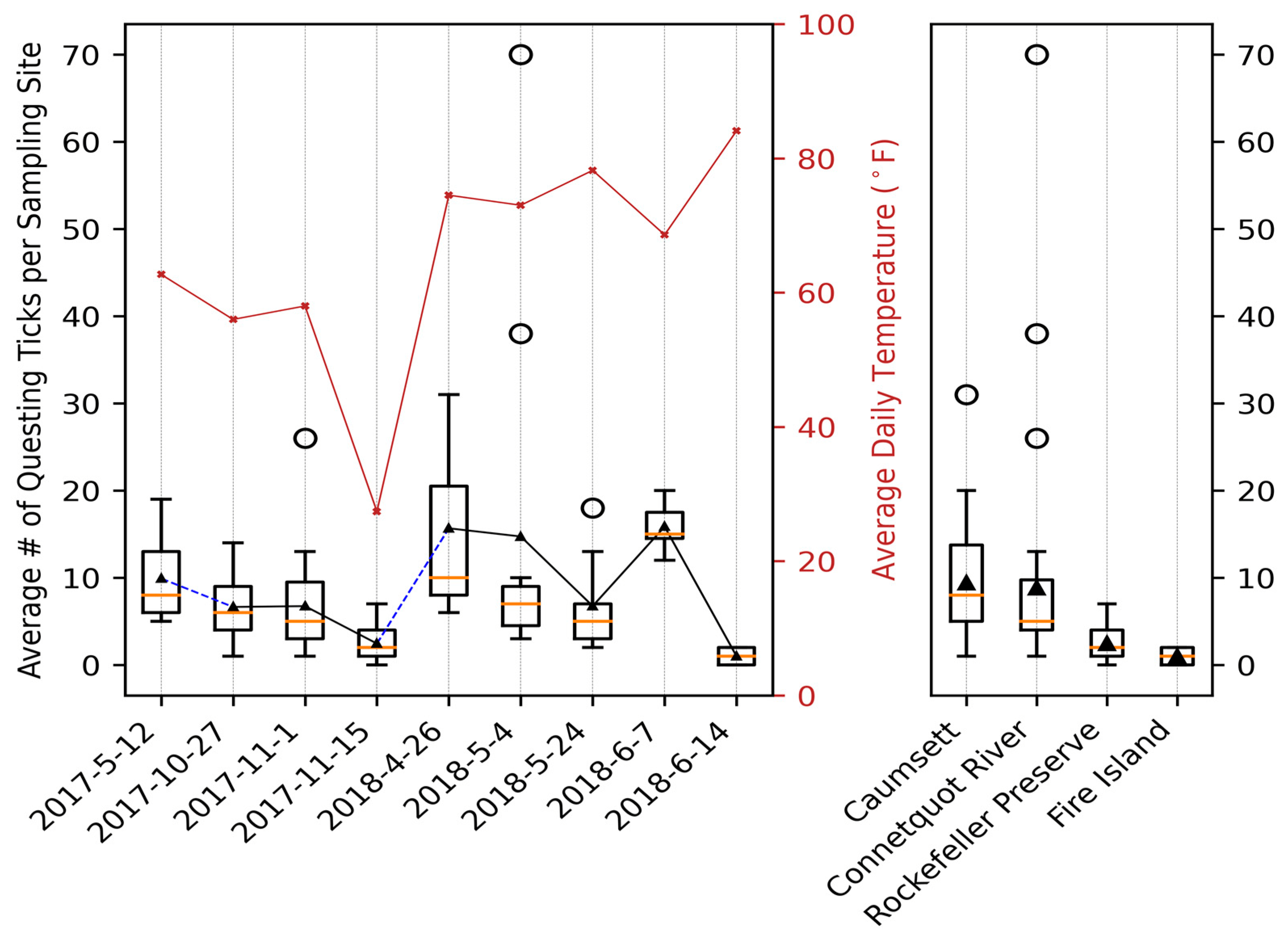
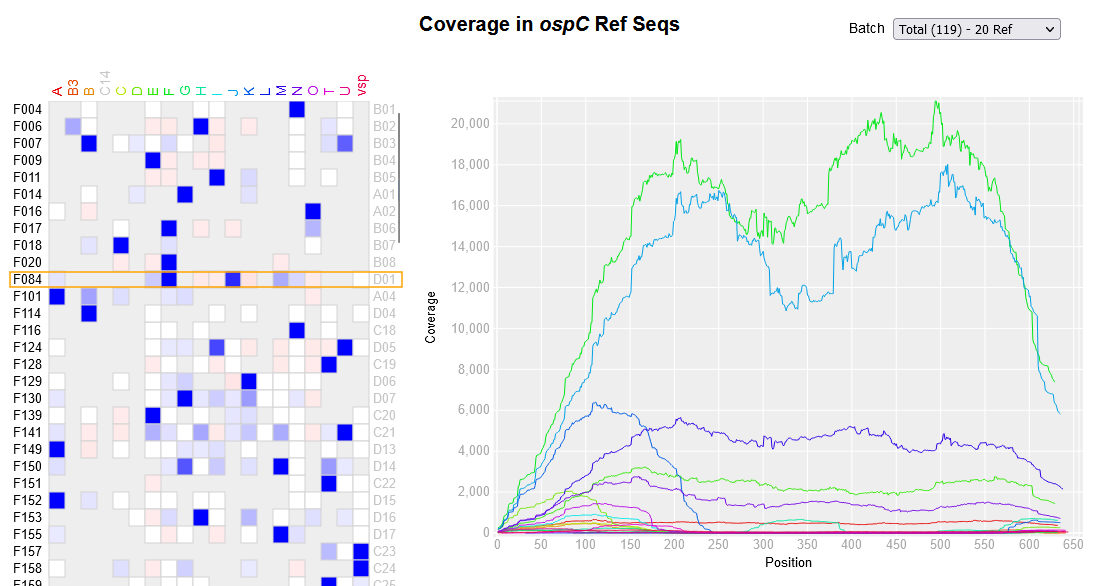
File:Applsci-13-11587-g004.png | link=https://doi.org/10.3390/app132011587 | Di, Chong, Brian Sulkow, Weigang Qiu, and Shipeng Sun. 2023. "Effects of Micro-Scale Environmental Factors on the Quantity of Questing Black-Legged Ticks in Suburban New York" '''''Applied Sciences''''' 13, no. 20: 11587. | File:Applsci-13-11587-g004.png | link=https://doi.org/10.3390/app132011587 | Di, Chong, Brian Sulkow, Weigang Qiu, and Shipeng Sun. 2023. "Effects of Micro-Scale Environmental Factors on the Quantity of Questing Black-Legged Ticks in Suburban New York" '''''Applied Sciences''''' 13, no. 20: 11587. | ||
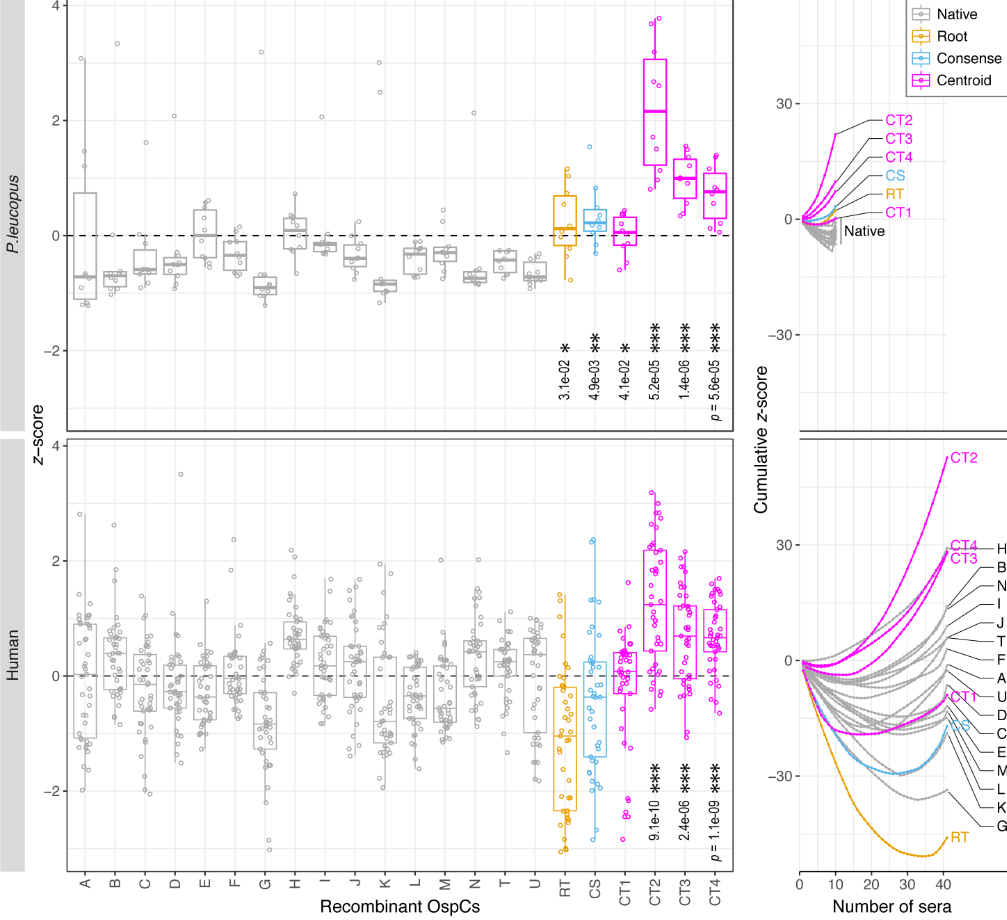
File:Spectrum.01743-22-f003.gif | link=https://doi.org/10.1128/spectrum.01743-22 | Li, Di, Zeglis, Qiu (2022). “Evolution of the ''vls'' antigenic variability locus of the Lyme Disease pathogen and development of recombinant monoclonal antibodies targeting conserved VlsE epitopes”. '''''Microbial Spectrum''''' 10 (5):1-15. | File:Spectrum.01743-22-f003.gif | link=https://doi.org/10.1128/spectrum.01743-22 | Li, Di, Zeglis, Qiu (2022). “Evolution of the ''vls'' antigenic variability locus of the Lyme Disease pathogen and development of recombinant monoclonal antibodies targeting conserved VlsE epitopes”. '''''Microbial Spectrum''''' 10 (5):1-15. | ||
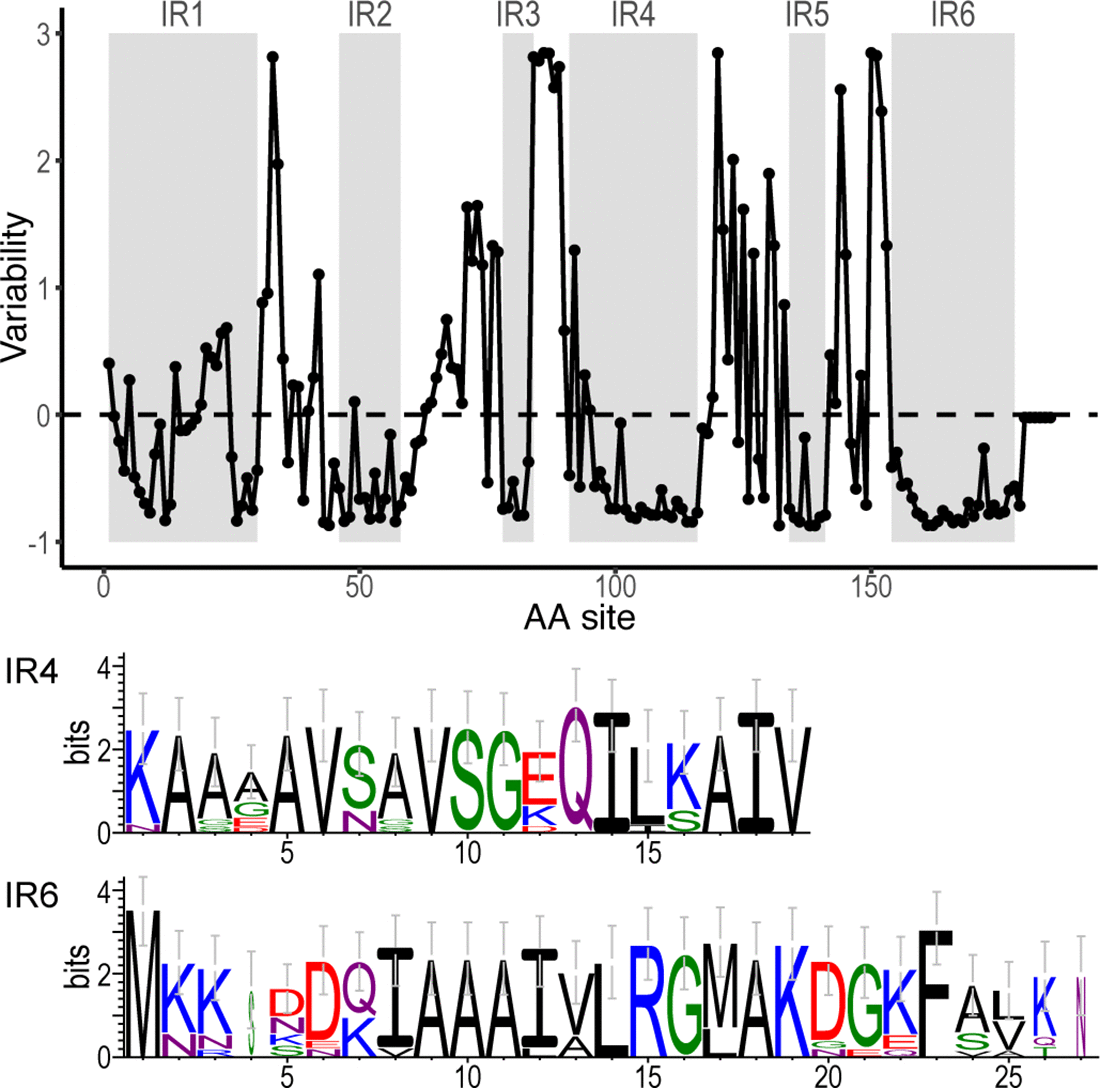
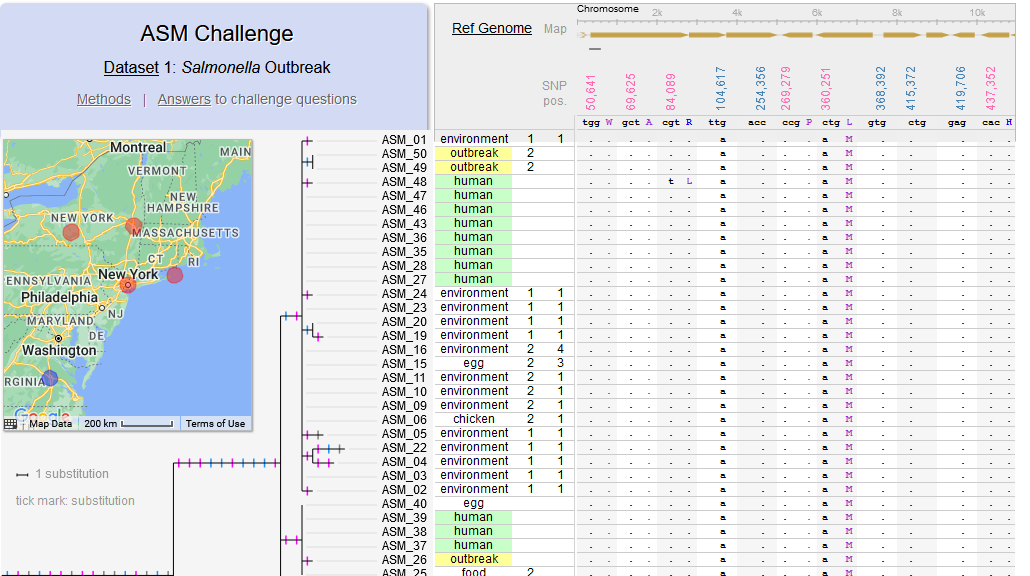
File:Fig7-small.png | link= https://pubmed.ncbi.nlm.nih.gov/34413477 | Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". '''''The ISME Journal'''''. 16, 447-464. (*co-first authors) [https:// | File:Fig7-small.png | link= https://pubmed.ncbi.nlm.nih.gov/34413477 | Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". '''''The ISME Journal'''''. 16, 447-464. (*co-first authors) [https://communities.springernature.com/posts/jenner-s-dilemma-and-how-to-win-evolutionary-arms-races-against-microbial-pathogens Blog Post] | ||
File:Ira-fig2.png | link=https://doi.org/10.21775/9781913652616 | Schwartz, Margos, Casjens, Qiu, Eggers (2020). "Multipartite Genome of Lyme Disease ''Borrelia'': Structure, Variation and Prophages". '''''Current Issues in Molecular Biology'''''. 42:409-454. | File:Ira-fig2.png | link=https://doi.org/10.21775/9781913652616 | Schwartz, Margos, Casjens, Qiu, Eggers (2020). "Multipartite Genome of Lyme Disease ''Borrelia'': Structure, Variation and Prophages". '''''Current Issues in Molecular Biology'''''. 42:409-454. | ||
| Line 58: | Line 63: | ||
(Fall 2022, Spring 2023 & Summer 2023) | (Fall 2022, Spring 2023 & Summer 2023) | ||
|| | || | ||
* Brandon Ely: CUNY Grad Center, Biology/MCD doctoral program | * Brandon Ely: CUNY Grad Center, Biology/MCD doctoral program | ||
* Dr Yozen Hernandez: System administrator (part-time), Ph.D. from Boston University | * Dr Yozen Hernandez: System administrator (part-time), Ph.D. from Boston University | ||
|| | || | ||
* Tasmina Hassan: Hunter Bio/CS | * Tasmina Hassan: Hunter Bio/CS | ||
| Line 67: | Line 70: | ||
* Tara Doma Lama: Hunter Bio/Bioinformatics | * Tara Doma Lama: Hunter Bio/Bioinformatics | ||
* Hagar Abuzaid: Hunter Bio | * Hagar Abuzaid: Hunter Bio | ||
|- | |- | ||
| Alumni (Since Fall 2002) | | Alumni (Since Fall 2002) | ||
|| | || | ||
* Dr Lia Di: Ph.D. from Nanjing Agricultural University & Wisconsin Blood Institute | * Li Li (Lily, 2023): CUNY Grad Center, Biology/EEB doctoral program | ||
* Dr Lia Di: Ph.D. Postdoctoral Research Associate, from Nanjing Agricultural University & Wisconsin Blood Institute | |||
* Dr Saymon Akther (2022): CUNY Grad Center, Biology/EEB | * Dr Saymon Akther (2022): CUNY Grad Center, Biology/EEB | ||
* Dr Rayees Rahman: Hunter Bio/Bioinformatics, Ph.D. from Mt Sinai Medical School | * Dr Rayees Rahman: Hunter Bio/Bioinformatics, Ph.D. from Mt Sinai Medical School | ||
| Line 81: | Line 84: | ||
* Dr Vincent Xue: CUNY CS/Bioinformatics, Ph.D. from MIT | * Dr Vincent Xue: CUNY CS/Bioinformatics, Ph.D. from MIT | ||
* Dr Fubin Li: CUNY Grad Center, Biology/MCD (Dr Laurel Eckhardt) | * Dr Fubin Li: CUNY Grad Center, Biology/MCD (Dr Laurel Eckhardt) | ||
* Dr Oliver Attie: Postdoctoral Research Associate, Ph.D. from NYU | |||
|| | || | ||
(published coauthors)<br> | (published coauthors)<br> | ||
| Line 104: | Line 108: | ||
==Web Apps== | ==Web Apps== | ||
===Apps with Collaborators=== | ===Apps with Collaborators (or from published papers)=== | ||
<gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | <gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | ||
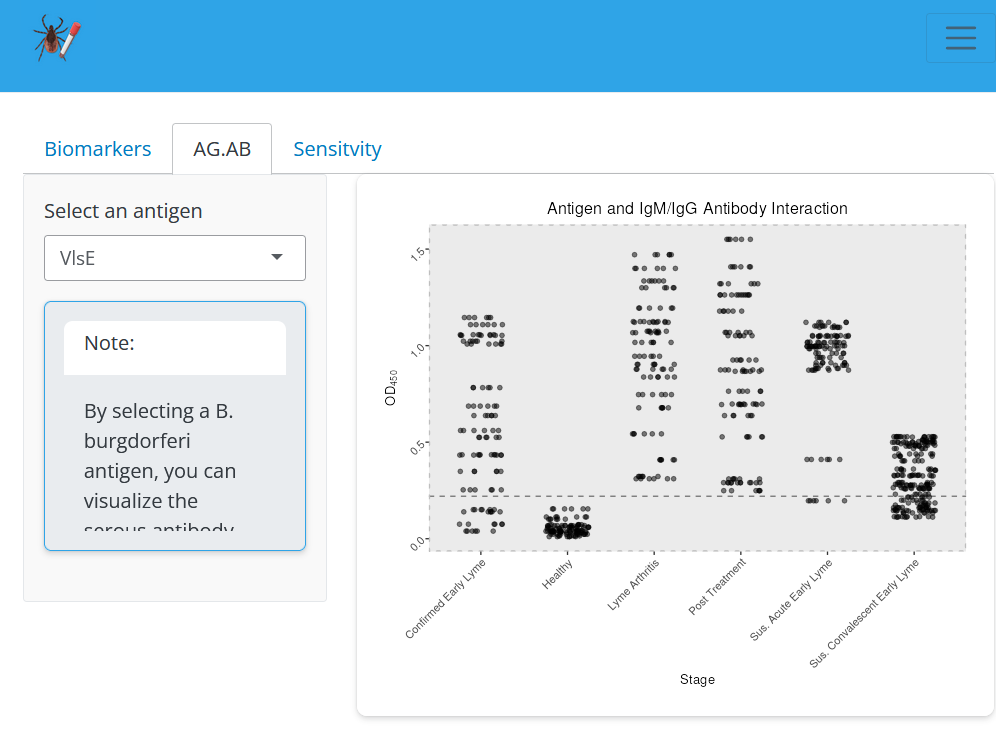
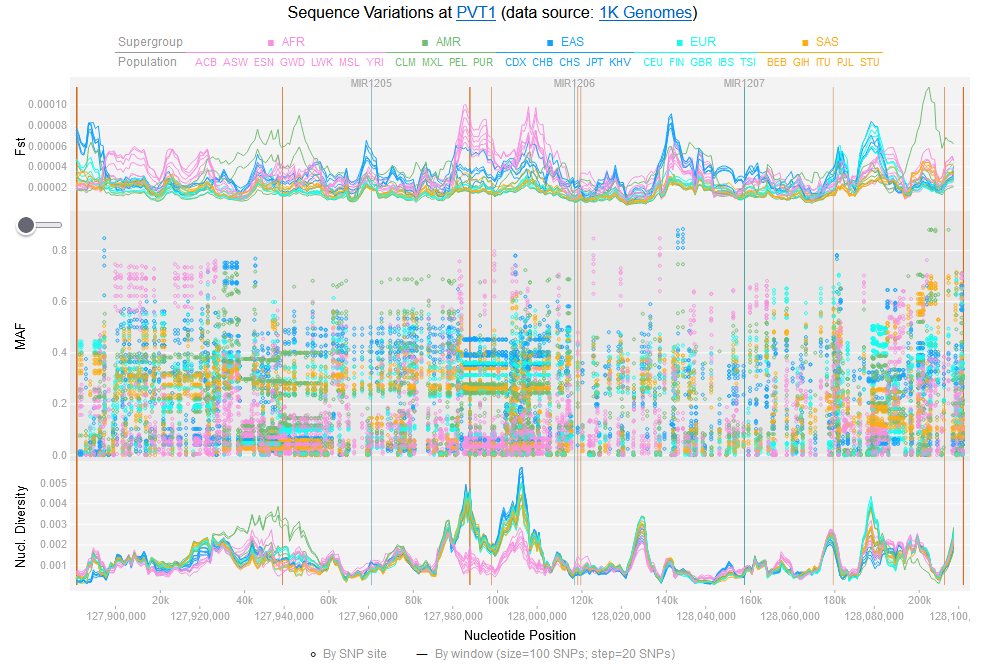
File:Screenshot_2024-06-19_205621.png | link=https://cov.genometracker.org/finalelisa-app/ | Borrelia diagnostic antigens (App developed by Liann Aris-Henry; Data from [https://journals.asm.org/doi/10.1128/jcm.01142-19 Arumugam et al (2019)]) | |||
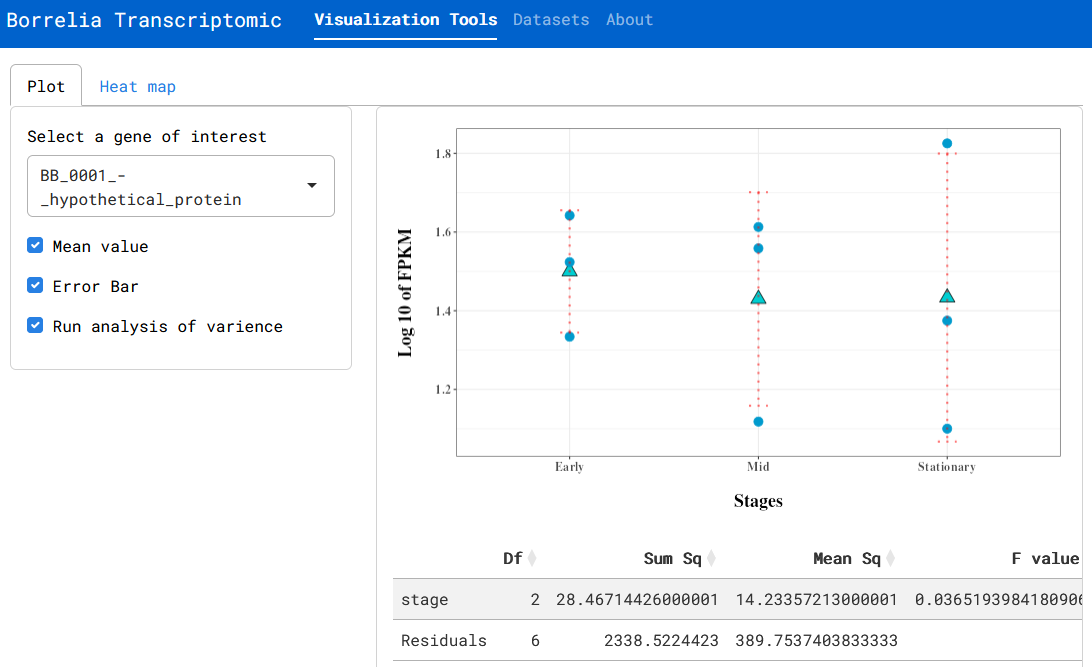
File:Screenshot_2024-06-18_144931.png | link=https://cov.genometracker.org/borrelia-app/ | Borrelia growth transcriptome (App developed by Laziz Asamov; Data from [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0164165 Arnold et al (2016)]) | |||
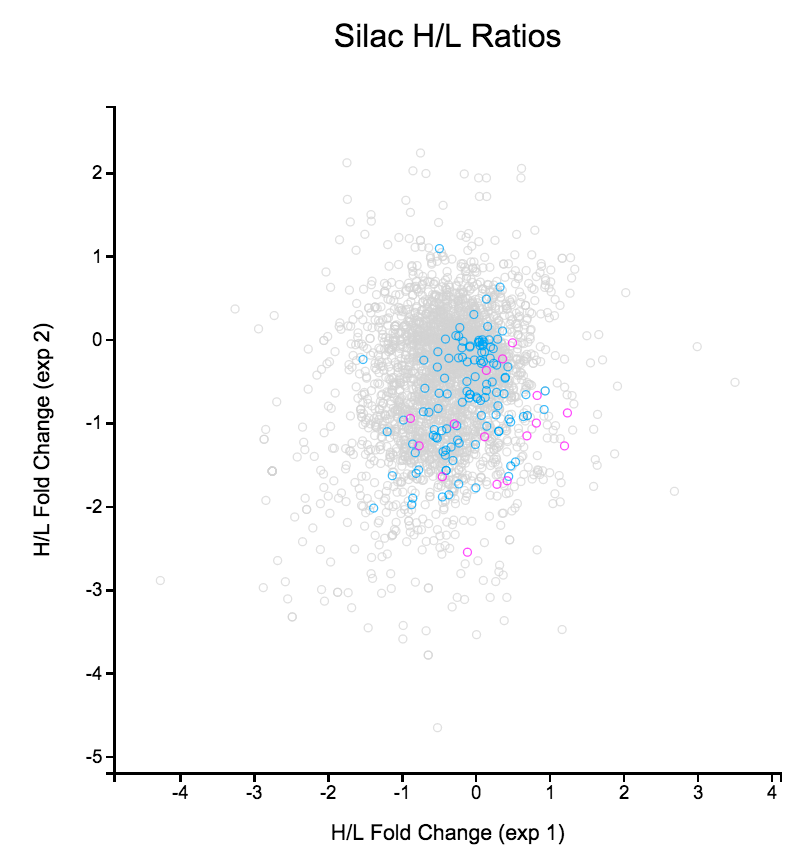
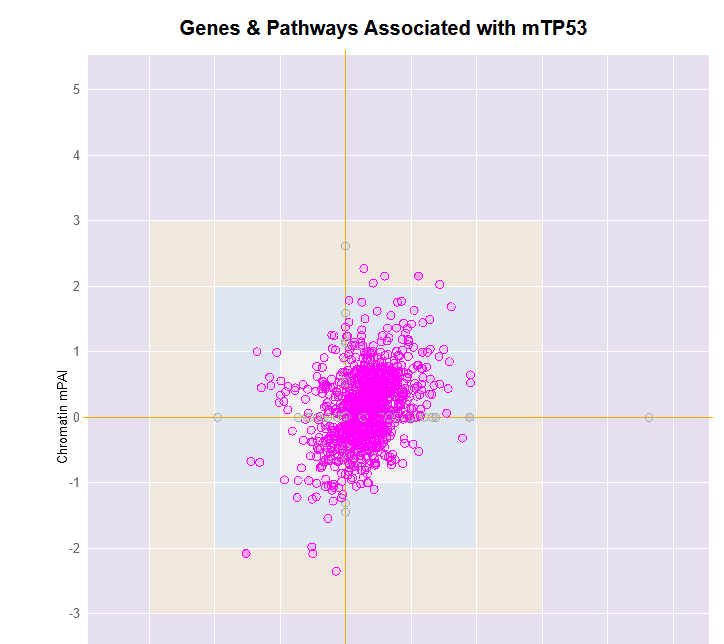
File: Silac.png | link=http://borreliabase.org/~wgqiu/Polotskaia_etal_2014/supp-fig-s1/ | Data & [http://borreliabase.org/~wgqiu/Polotskaia_etal_2014/supp-table-s1/ GSEA] associated with Polotskaia_etal_2014 (with Bargonetti Lab @Hunter) | File: Silac.png | link=http://borreliabase.org/~wgqiu/Polotskaia_etal_2014/supp-fig-s1/ | Data & [http://borreliabase.org/~wgqiu/Polotskaia_etal_2014/supp-table-s1/ GSEA] associated with Polotskaia_etal_2014 (with Bargonetti Lab @Hunter) | ||
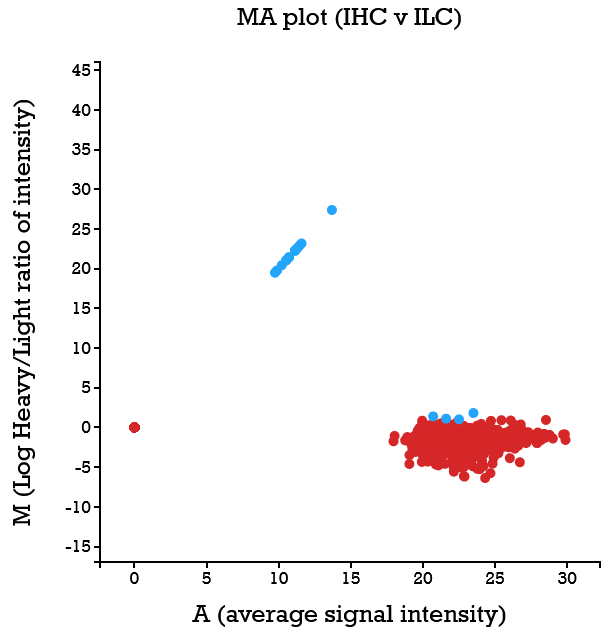
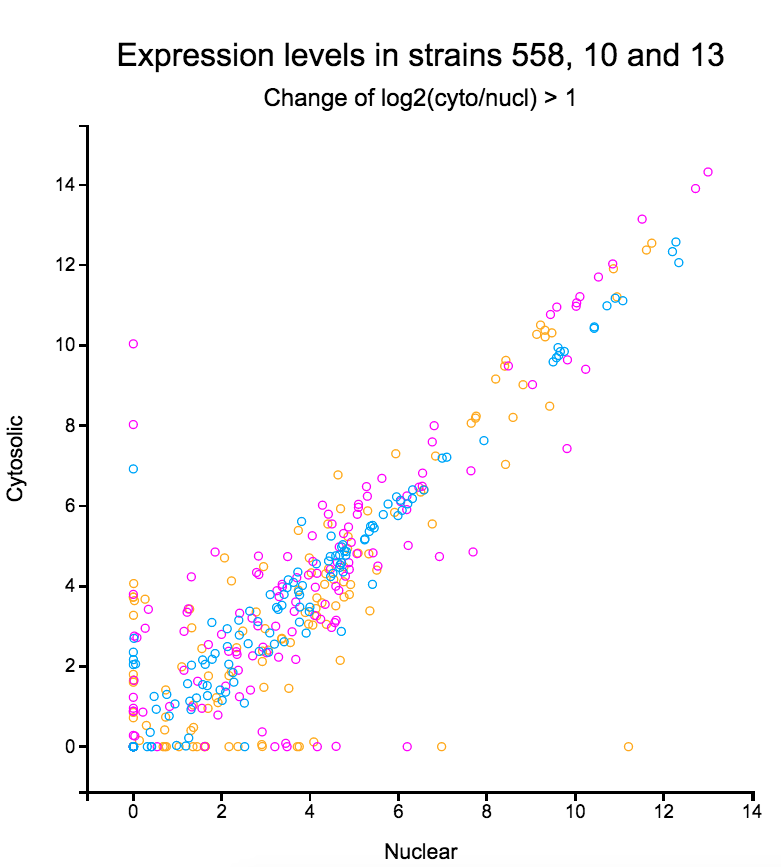
File:Screenshot_2023-03-24_122016.png | link=http://borreliabase.org/~wgqiu/clickme-khalikuz/temp-Points.html | Phosphoproteome MDM2KD data set (with Bargonetti Lab @Hunter) | File:Screenshot_2023-03-24_122016.png | link=http://borreliabase.org/~wgqiu/clickme-khalikuz/temp-Points.html | Phosphoproteome MDM2KD data set (with Bargonetti Lab @Hunter) | ||
| Line 128: | Line 134: | ||
</gallery> | </gallery> | ||
== | ==Curricular Development & Bioinformatics/QuBi Advising== | ||
* QuBi advisors: Weigang Qiu, Ntino Krampis, Rabindra Mandal (Biology); Saad Mneimeih, Lei Xie (CS); Akira Kawamura (Chem); Dana Sylvan (Math & Stats) | * QuBi advisors: Weigang Qiu, Ntino Krampis, Rabindra Mandal (Biology); Saad Mneimeih, Lei Xie (CS); Akira Kawamura (Chem); Dana Sylvan (Math & Stats) | ||
** Permission for non-Biology majors to take BIOL203 & BIOL425, every Spring | ** Permission for non-Biology majors to take BIOL203 & BIOL425, every Spring | ||
| Line 148: | Line 153: | ||
* General advising: | * General advising: | ||
** ~40 students every semester. Send out emails to students. Go through student courses by Email or by appointment | ** ~40 students every semester. Send out emails to students. Go through student courses by Email or by appointment | ||
** Recommend new math courses: MATH15200 & STAT21350 | ** Recommend new math courses: '''MATH15200 & STAT21350''' | ||
* Hosting QuBi students in lab | * Hosting QuBi students in lab | ||
** This is to enhance the informatics and coding skills of our students | ** This is to enhance the informatics and coding skills of our students | ||
** Students should register and get BIOL48002 credits, which counts towards their elective credits & eligibility for honors | ** Students should register and get '''BIOL48002''' credits, which counts towards their elective credits & eligibility for honors | ||
** 3-5 students per semester | ** 3-5 students per semester | ||
* Outside research opportunities | * Outside research opportunities | ||
| Line 157: | Line 162: | ||
** Simons Foundation/Flatiron Institute Center for Computational Biology (CCB) Internship program. Open House in Spring | ** Simons Foundation/Flatiron Institute Center for Computational Biology (CCB) Internship program. Open House in Spring | ||
==Course/Lecture syllabi== | |||
* [[NYRaMP-Informatics-2025|NYRaMP Workshop (August 2025, by Brandon Ely)]] | |||
* [[Computational Genomics (KIZ, Fall 2024)]] | |||
* [[NYRaMP-Informatics-2024|NYRaMP Workshop (August 2024)]] | |||
* BIOL47120 BioMedical Genomics (Spring 2024). Tutorials: [https://borreliabase.org/~wgqiu/tutorial-markdown.html R Markdown] [https://borreliabase.org/~wgqiu/cluster-analysis.html Cluster analysis] [https://borreliabase.org/~wgqiu/scRNA-analysis.html single-cell RNA-seq] | |||
* BIOL425 Computational Molecular Biology (Spring, 2023). [https://github.com/weigangq/CSB-BIOL425/tree/master/lecture-materials Lecture material on github] | * BIOL425 Computational Molecular Biology (Spring, 2023). [https://github.com/weigangq/CSB-BIOL425/tree/master/lecture-materials Lecture material on github] | ||
* [http://borreliabase.org/~wgqiu/r-demo-2023.html | * BIOL714 Cell Biology: [http://borreliabase.org/~wgqiu/r-demo-2024.html R Demo (Spring 2024)] [http://borreliabase.org/~wgqiu/r-demo-2023.html R Demo (Spring 2023)] | ||
* QuBi module: [[QuBi/module/bio203-lab12—2022|BIOL20300 Molecular Genetics, Lab 12 (2023)]] | * QuBi module: [[QuBi/module/bio203-lab12—2022|BIOL20300 Molecular Genetics, Lab 12 (2023)]] | ||
* QuBi module: [[QuBi/modules/biol203-geno-pheno-association-2022|BIOL20300 Molecular Genetics, Lab 13 (2022)]] | * QuBi module: [[QuBi/modules/biol203-geno-pheno-association-2022|BIOL20300 Molecular Genetics, Lab 13 (2022)]] | ||
* QuBi module: [[QuBi/modules/biol303|BIOL30300 Cell Biology, Bioinformatics Lab (transcriptome analysis)]] | * QuBi module: [[QuBi/modules/biol303|BIOL30300 Cell Biology, Bioinformatics Lab (transcriptome analysis)]] | ||
* [[BigData 2020|Big Data (Summer, 2020)]] | * [[BigData 2020|Big Data (Summer, 2020)]] | ||
* [[BioMed-R-2020|BIOL47120 Biomedical Genomics II (Spring, 2020)]] | * [[BioMed-R-2020|BIOL47120 Biomedical Genomics II (Spring, 2020)]] | ||
** [http://borreliabase.org/~wgqiu/tutorial-markdown.html Tutorial: R Markdown (Spring 2024)] | |||
** [http://borreliabase.org/~wgqiu/cluster-analysis.html Tutorial: Cluster analysis (Spring 2024)] | |||
** [http://borreliabase.org/~wgqiu/scRNA-analysis.html Tutorial: single-cel transcriptome analysis (Spring 2024)] | |||
* [[Biol425 2020|BIOL425 Computational Molecular Biology (Spring, 2020)]] | * [[Biol425 2020|BIOL425 Computational Molecular Biology (Spring, 2020)]] | ||
* [[Biol375 2019|BIOL37500, Molecular Evolution (Fall, 2019)]] | * [[Biol375 2019|BIOL37500, Molecular Evolution (Fall, 2019)]] | ||
| Line 173: | Line 185: | ||
==SARS-CoV-2 genome evolution== | ==SARS-CoV-2 genome evolution== | ||
<gallery mode="packed" heights="200px" perrow="3" style="text-align:left"> | <gallery mode="packed" heights="200px" perrow="3" style="text-align:left"> | ||
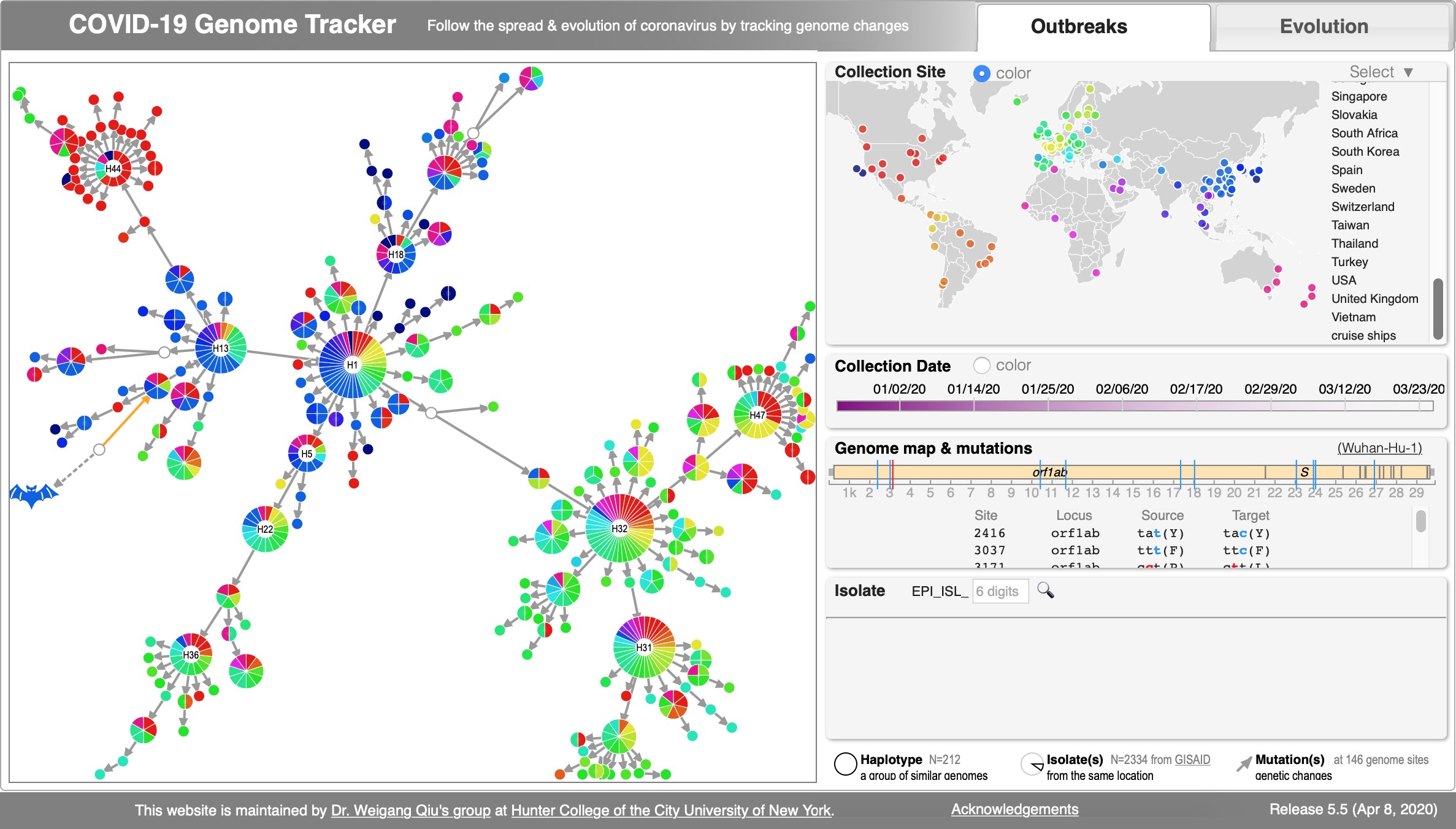
File:Cov-fig1.jpg | Akther, Bezrucenkovas, Sulkow, Panlasigui, Qiu, Di (April, 2020). "CoV Genome Tracker: tracing genomic footprints of Covid-19 pandemic". '''''[https://www.biorxiv.org/content/biorxiv/early/2020/04/14/2020.04.10.036343.full.pdf BioRxiv]'''''; Web app: SARS-CoV-2 Genome Tracker; Github: https://github.com/weigangq/cov-browser | File:Cov-fig1.jpg | Akther, Bezrucenkovas, Sulkow, Panlasigui, Qiu, Di (April, 2020). "CoV Genome Tracker: tracing genomic footprints of Covid-19 pandemic". '''''[https://www.biorxiv.org/content/biorxiv/early/2020/04/14/2020.04.10.036343.full.pdf BioRxiv]'''''; [https://cov.genometracker.org/ Web app: SARS-CoV-2 Genome Tracker]; Github: https://github.com/weigangq/cov-browser | ||
File:Rec-fig2.png | Akther, Li, Martin, Di, Sulkow, Pante, Bezrucenlovas, Luft, Qiu (May, 2020). "Origin, recombination, and missed opprotunities: a genomic perspective of the first 100 days of COVID-19 pandemic". (Unpublished). | File:Rec-fig2.png | Akther, Li, Martin, Di, Sulkow, Pante, Bezrucenlovas, Luft, Qiu (May, 2020). "Origin, recombination, and missed opprotunities: a genomic perspective of the first 100 days of COVID-19 pandemic". (Unpublished). | ||
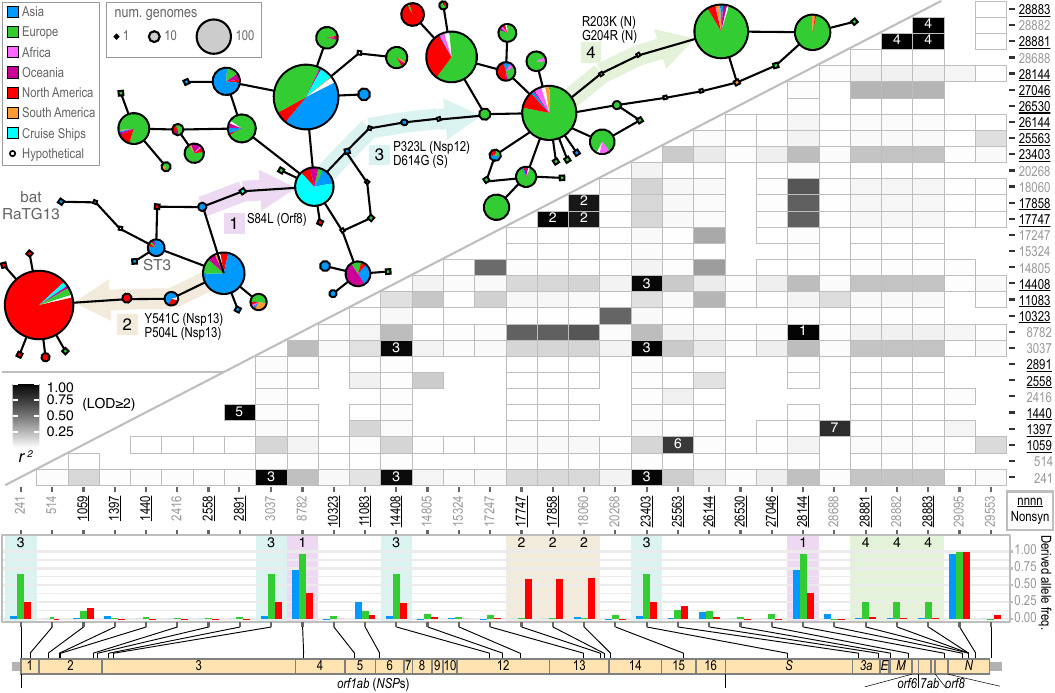
File:Cov-fig3-trace.png | Akther, Bezrucenlovas, Li, Sulkow, Di, Pante, Martin, Luft, Qiu (Sep, 2021). "Following the Trail of One Million Genomes: Footprints of SARS-CoV-2 Adaptation to Humans". '''''[https://www.biorxiv.org/content/biorxiv/early/2021/05/10/2021.05.07.443114.full.pdf BioRxiv link]''''': . Github: https://github.com/weigangq/cov-db | File:Cov-fig3-trace.png | Akther, Bezrucenlovas, Li, Sulkow, Di, Pante, Martin, Luft, Qiu (Sep, 2021). "Following the Trail of One Million Genomes: Footprints of SARS-CoV-2 Adaptation to Humans". '''''[https://www.biorxiv.org/content/biorxiv/early/2021/05/10/2021.05.07.443114.full.pdf BioRxiv link]''''': . Github: https://github.com/weigangq/cov-db | ||
| Line 179: | Line 191: | ||
==Lab Resources & Protocols== | ==Lab Resources & Protocols== | ||
* Nanopore DNA barcoding protocol: https://nanopore4edu.org/latest/annotated_experiments/dna_barcoding/ | |||
* [[Monte Carlo Club]] | |||
* [[NY-RaMP Mentoring]] | |||
* Borreliella genome sequencing consortium: Weekly meetings (Tu @11): Since Jan 2023 | * Borreliella genome sequencing consortium: Weekly meetings (Tu @11): Since Jan 2023 | ||
* Borreliella diagnostic antigens (Fall 2023-Fall 2027): | * Borreliella diagnostic antigens (Fall 2023-Fall 2027): | ||
| Line 186: | Line 201: | ||
* Qiu lab Github repositories: https://github.com/weigangq/?tab=repositories | * Qiu lab Github repositories: https://github.com/weigangq/?tab=repositories | ||
* [[Mini-Tutorals|Mini-Protocols]] (frequently used computer codes and pipelines) | * [[Mini-Tutorals|Mini-Protocols]] (frequently used computer codes and pipelines) | ||
* | * Python tutorial: https://wiki.genometracker.org/~weigang/Intro_to_Python.html | ||
* | * [[Tick protocol|ick handling protocols]] | ||
* [[A Primer on the Cluster System at Hunter|Hunter HPC Usage]] | * [[A Primer on the Cluster System at Hunter|Hunter HPC Usage]] | ||
* [https://r4ds.hadley.nz/ R for Data Science (2e)], (2024) by Wickham, Grolemund & Çetinkaya-Rundel ([https://bookdown.org/ Bookdown version]) | * [https://r4ds.hadley.nz/ R for Data Science (2e)], (2024) by Wickham, Grolemund & Çetinkaya-Rundel ([https://bookdown.org/ Bookdown version]) | ||
* Borrelia Genome Consortium: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA431102/ | |||
* Canadian Bbsl genome assemblies: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1130942 | |||
* Nanopore sequencing resources: | |||
** eBook: https://store.nanoporetech.com/us/minion.html | |||
== Wiki Help == | == Wiki Help == | ||
Latest revision as of 20:53, 28 October 2025
Weigang Qiu, Ph.D., Professor, Department of Biological Sciences Hunter College of City University of New York 413 East 69th Street, New York, NY 10021 Office: 1-212-896-0445 Email: wqiu-at-(hunter.cuny.edu) |
Fieldwork Gallery
Lab publications
Lyme Genomics, Evolution, & Ecology
Akther et al. 2024. "Natural selection and recombination at host-interacting lipoprotein loci drive genome diversification of Lyme disease and related bacteria" mBio 0:e01749-24. Press coverage: CUNY Graduate Center News Story; Hunter press
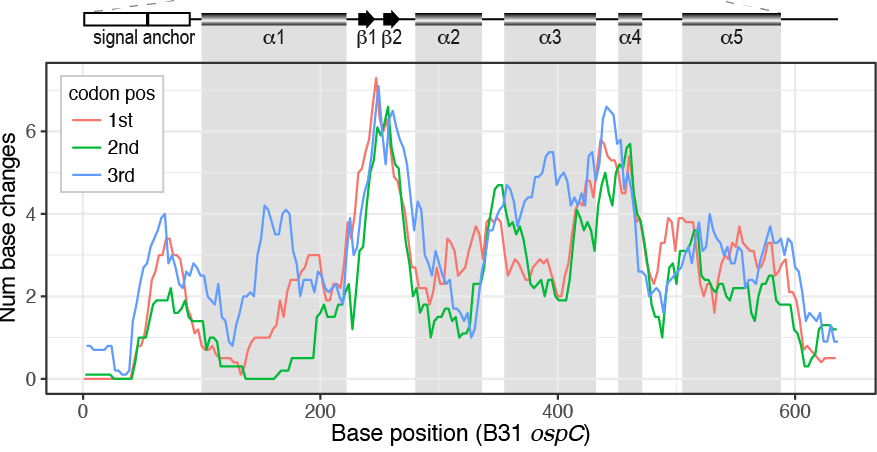
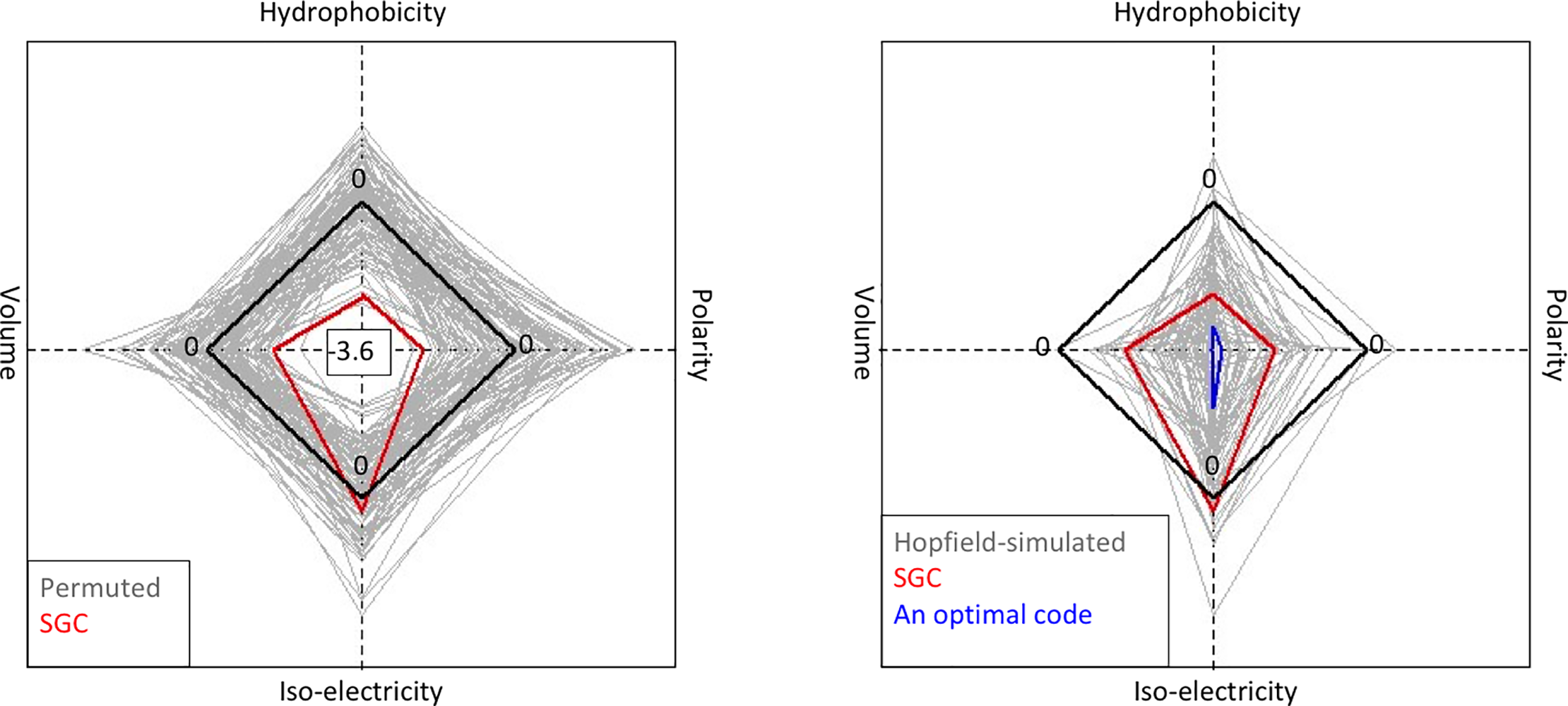
Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". The ISME Journal. 16, 447-464. (*co-first authors) Blog Post
Evolution & Learning Algorithms
Informatics Tool Development
last update: March 20, 2023
Lab members and trainees
| Year/Period | Doctoral members & trainees | Other members & trainees |
|---|---|---|
| Current Academic Year
(Fall 2022, Spring 2023 & Summer 2023) |
|
|
| Alumni (Since Fall 2002) |
|
(published coauthors)
|
Web Apps
Apps with Collaborators (or from published papers)
Borrelia diagnostic antigens (App developed by Liann Aris-Henry; Data from Arumugam et al (2019))
Borrelia growth transcriptome (App developed by Laziz Asamov; Data from Arnold et al (2016))
Data & GSEA associated with Polotskaia_etal_2014 (with Bargonetti Lab @Hunter)
Qiu Lab Apps
Curricular Development & Bioinformatics/QuBi Advising
- QuBi advisors: Weigang Qiu, Ntino Krampis, Rabindra Mandal (Biology); Saad Mneimeih, Lei Xie (CS); Akira Kawamura (Chem); Dana Sylvan (Math & Stats)
- Permission for non-Biology majors to take BIOL203 & BIOL425, every Spring
- Collect names, major, and IDs to send to course coordinator to grant permission. Waive BIOL10200 pre-reqs for taking BIOL203.
- Curricular resources:
- Biology courses and pre-reqs
- Hunter Biology Major 1 (including the Bioinformatics Option)
- Hunter Chemistry Major 2 (including the Bioinformatics Option)
- Hunter Computer Science (including Bioinformatics Concentration)
- Hunter Mathematics (including the Quantitative Biology Concentration)
- Hunter Statistics (including the Quantitative Biology Concentration)
- QuBi advising:
- Declaration of Bioinformatics concentration: In-person advising to work out the semester-by-semester courses
- Approve on department spreadsheet (or send email to "Samantha Sheppard-Lahiji" and "HTR Bio" <biology@hunter.cuny.edu>)
- Students should take Bioinformatics-specific electives (8 cred; see Hunter Catalog below), not general electives
- Examples: Anthrop302 (3 cr); Chem333 (3 cr); BIOL47119 & BIOL47120 (3cr); BIOL48002 (2 cr)
- Students need to take BIOL48002 (2 cr), which counts towards as research credit, to graduate as honors
- General advising:
- ~40 students every semester. Send out emails to students. Go through student courses by Email or by appointment
- Recommend new math courses: MATH15200 & STAT21350
- Hosting QuBi students in lab
- This is to enhance the informatics and coding skills of our students
- Students should register and get BIOL48002 credits, which counts towards their elective credits & eligibility for honors
- 3-5 students per semester
- Outside research opportunities
- MIT Quantitative Workshop (first week of January, in Boston). Coordination with CS (Saad & Susan Epstein) in Fall
- Simons Foundation/Flatiron Institute Center for Computational Biology (CCB) Internship program. Open House in Spring
Course/Lecture syllabi
- NYRaMP Workshop (August 2025, by Brandon Ely)
- Computational Genomics (KIZ, Fall 2024)
- NYRaMP Workshop (August 2024)
- BIOL47120 BioMedical Genomics (Spring 2024). Tutorials: R Markdown Cluster analysis single-cell RNA-seq
- BIOL425 Computational Molecular Biology (Spring, 2023). Lecture material on github
- BIOL714 Cell Biology: R Demo (Spring 2024) R Demo (Spring 2023)
- QuBi module: BIOL20300 Molecular Genetics, Lab 12 (2023)
- QuBi module: BIOL20300 Molecular Genetics, Lab 13 (2022)
- QuBi module: BIOL30300 Cell Biology, Bioinformatics Lab (transcriptome analysis)
- Big Data (Summer, 2020)
- BIOL47120 Biomedical Genomics II (Spring, 2020)
- BIOL425 Computational Molecular Biology (Spring, 2020)
- BIOL37500, Molecular Evolution (Fall, 2019)
- Southwest University R course (Summer, 2019)
- Analysis of Biological Data (Spring, 2017)
- Bioinformatics Workshop (Summer, 2014)
SARS-CoV-2 genome evolution
Akther, Bezrucenkovas, Sulkow, Panlasigui, Qiu, Di (April, 2020). "CoV Genome Tracker: tracing genomic footprints of Covid-19 pandemic". BioRxiv; Web app: SARS-CoV-2 Genome Tracker; Github: https://github.com/weigangq/cov-browser
Akther, Bezrucenlovas, Li, Sulkow, Di, Pante, Martin, Luft, Qiu (Sep, 2021). "Following the Trail of One Million Genomes: Footprints of SARS-CoV-2 Adaptation to Humans". BioRxiv link: . Github: https://github.com/weigangq/cov-db
Lab Resources & Protocols
- Nanopore DNA barcoding protocol: https://nanopore4edu.org/latest/annotated_experiments/dna_barcoding/
- Monte Carlo Club
- NY-RaMP Mentoring
- Borreliella genome sequencing consortium: Weekly meetings (Tu @11): Since Jan 2023
- Borreliella diagnostic antigens (Fall 2023-Fall 2027):
- Zoom call (Jan 23, 2024)
- Next meeting: March 23, 2024
- Qiu lab network first-time user guide
- Qiu lab Github repositories: https://github.com/weigangq/?tab=repositories
- Mini-Protocols (frequently used computer codes and pipelines)
- Python tutorial: https://wiki.genometracker.org/~weigang/Intro_to_Python.html
- ick handling protocols
- Hunter HPC Usage
- R for Data Science (2e), (2024) by Wickham, Grolemund & Çetinkaya-Rundel (Bookdown version)
- Borrelia Genome Consortium: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA431102/
- Canadian Bbsl genome assemblies: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1130942
- Nanopore sequencing resources:
Wiki Help
- Configuration settings list
- MediaWiki FAQ
- MediaWiki release mailing list
- Localise MediaWiki for your language
- Learn how to combat spam on your wiki
- Consult the User's Guide for information on using the wiki software.