Main Page: Difference between revisions
Jump to navigation
Jump to search
imported>Weigang |
|||
| (293 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
< | {| | ||
< | |- | ||
| | |||
[[File:Protrait-July-2024-Qiu.jpg| x250 px | thumb]] | |||
| <center> <strong>Welcome to Qiu Lab Wiki @ Hunter</strong><br>Weigang Qiu, Ph.D., Professor, [http://biology.hunter.cuny.edu Department of Biological Sciences]<br> | |||
[https://hunter.cuny.edu Hunter College] of [https://www.cuny.edu City University of New York]<br> | |||
Adjunct Faculty, Weill Cornell Medical College, Department of Physiology and Biophysics<br> | |||
Belfer Research Building, Room 402<br>413 East 69th Street, New York, NY 10021<br>Office: 1-212-896-0445<br>Email: wqiu-at-(hunter.cuny.edu)<br> </center> | |||
== | | [[File:Belfer_building.jpg | x300px | thumb | [https://goo.gl/maps/xv1KmaW3XEnxaY1V7 Directions by Google Map]]] | ||
|__TOC__ | |||
<gallery> | |} | ||
==Fieldwork Gallery== | |||
<gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | |||
File:Tick-trip-2023.jpg|thumb|Oct 2023, Long Island, NY | |||
File:Tick-trip-2022.jpg|thumb|June 2022, MA | |||
File:Tick-trip-2021.jpg|thumb|June 2021, MA | |||
</gallery> | </gallery> | ||
<youtube width="200" height="150">Jb4ACK-GjM0</youtube> | |||
==Lab publications== | |||
===Lyme Genomics, Evolution, & Ecology=== | |||
<gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | |||
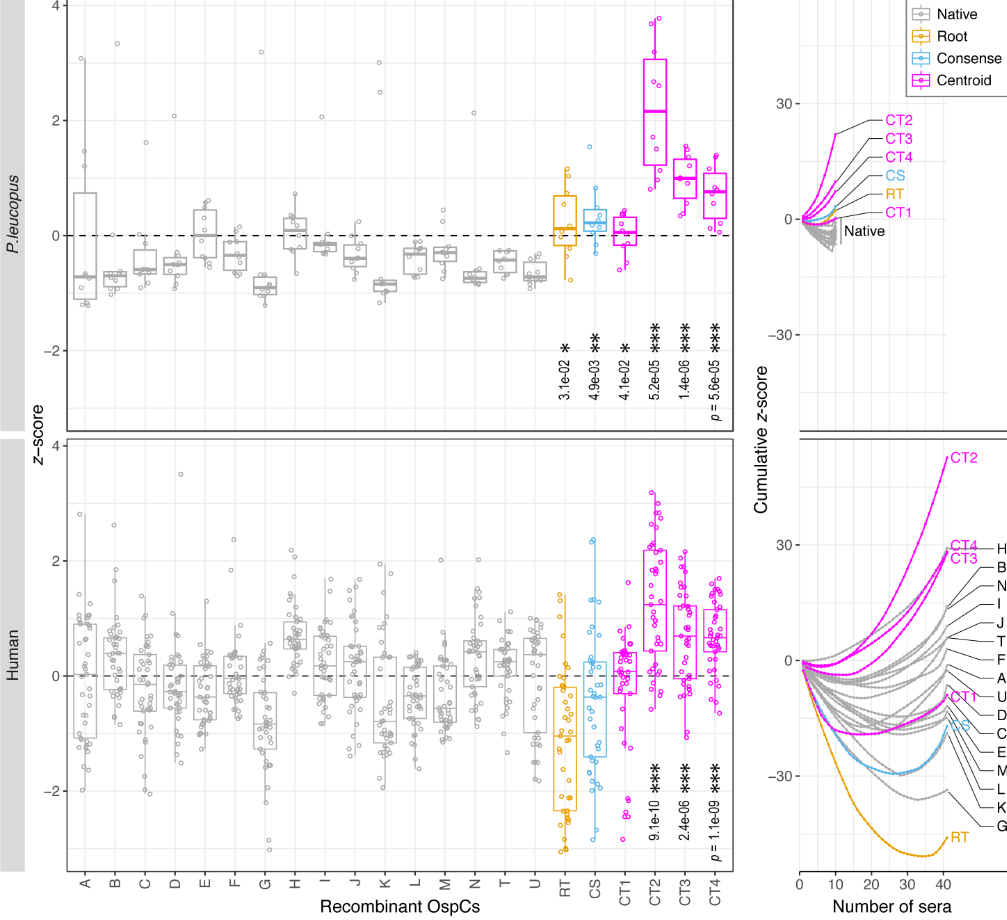
File:Screenshot_2024-08-15_094908.png | link=https://journals.asm.org/doi/10.1128/mbio.01749-24 | Akther et al. 2024. "Natural selection and recombination at host-interacting lipoprotein loci drive genome diversification of Lyme disease and related bacteria" '''''mBio''''' 0:e01749-24. Press coverage: [https://www.gc.cuny.edu/news/cuny-graduate-center-biologists-map-dna-lyme-disease-bacteria CUNY Graduate Center News Story]; [https://hunter.cuny.edu/news/hunter-researcher-maps-dna-of-lyme-disease-bacteria/ Hunter press] | |||
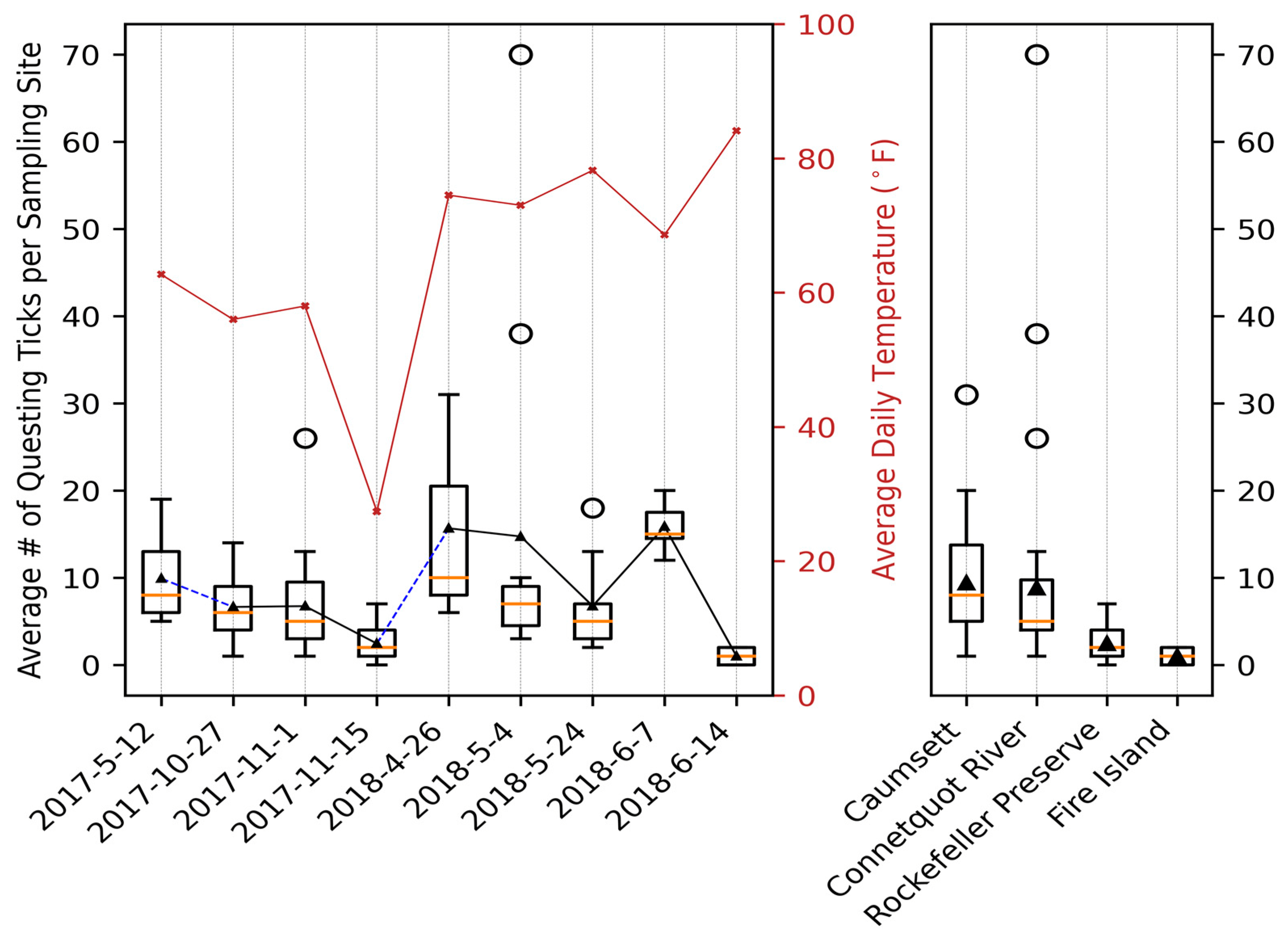
File:Applsci-13-11587-g004.png | link=https://doi.org/10.3390/app132011587 | Di, Chong, Brian Sulkow, Weigang Qiu, and Shipeng Sun. 2023. "Effects of Micro-Scale Environmental Factors on the Quantity of Questing Black-Legged Ticks in Suburban New York" '''''Applied Sciences''''' 13, no. 20: 11587. | |||
File:Spectrum.01743-22-f003.gif | link=https://doi.org/10.1128/spectrum.01743-22 | Li, Di, Zeglis, Qiu (2022). “Evolution of the ''vls'' antigenic variability locus of the Lyme Disease pathogen and development of recombinant monoclonal antibodies targeting conserved VlsE epitopes”. '''''Microbial Spectrum''''' 10 (5):1-15. | |||
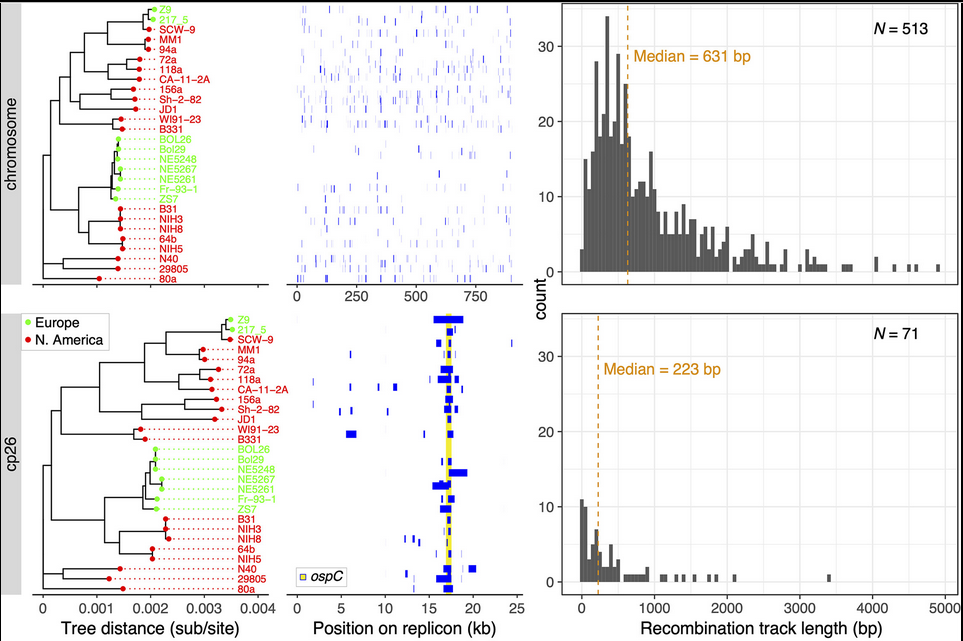
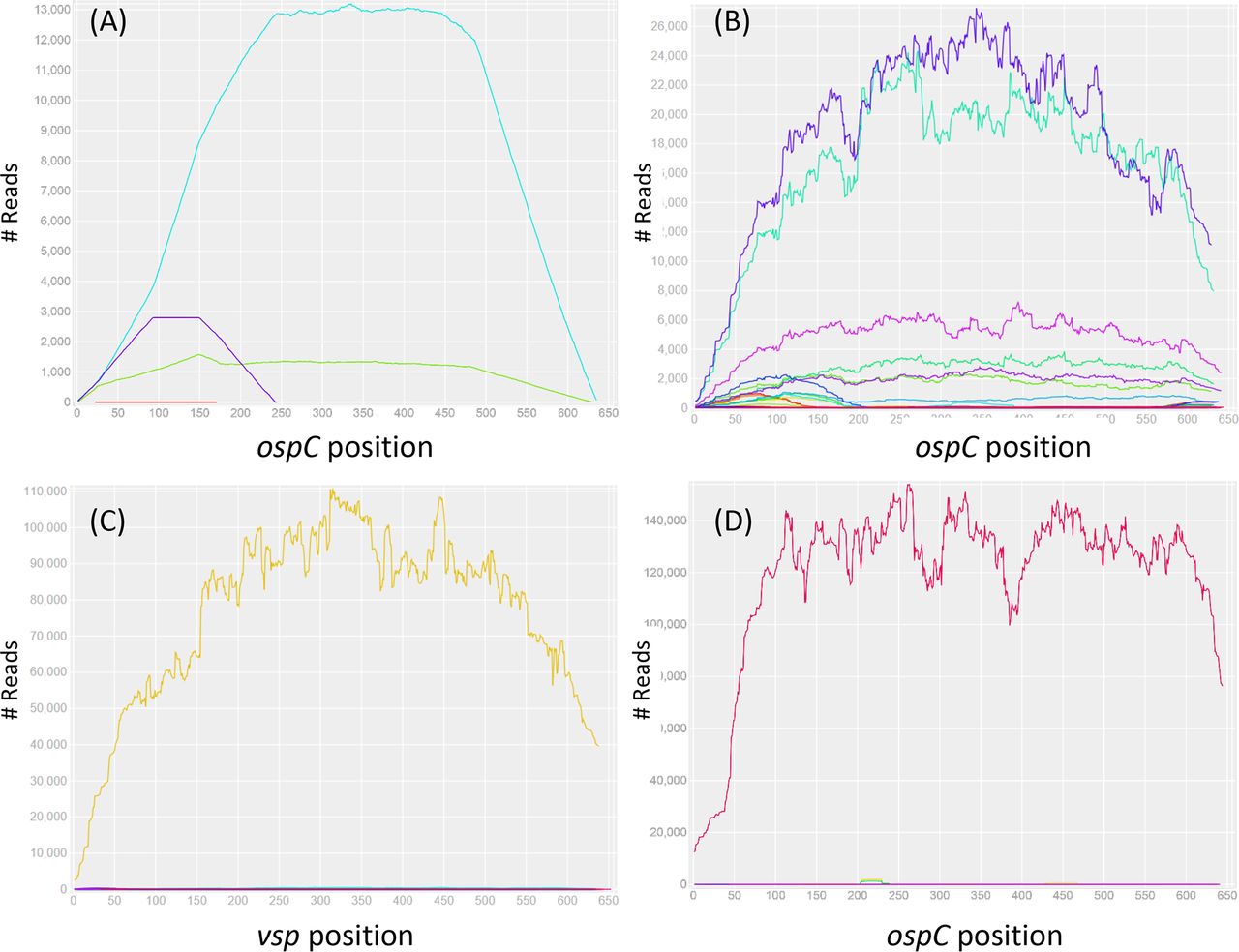
---- | File:Fig7-small.png | link= https://pubmed.ncbi.nlm.nih.gov/34413477 | Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". '''''The ISME Journal'''''. 16, 447-464. (*co-first authors) [https://communities.springernature.com/posts/jenner-s-dilemma-and-how-to-win-evolutionary-arms-races-against-microbial-pathogens Blog Post] | ||
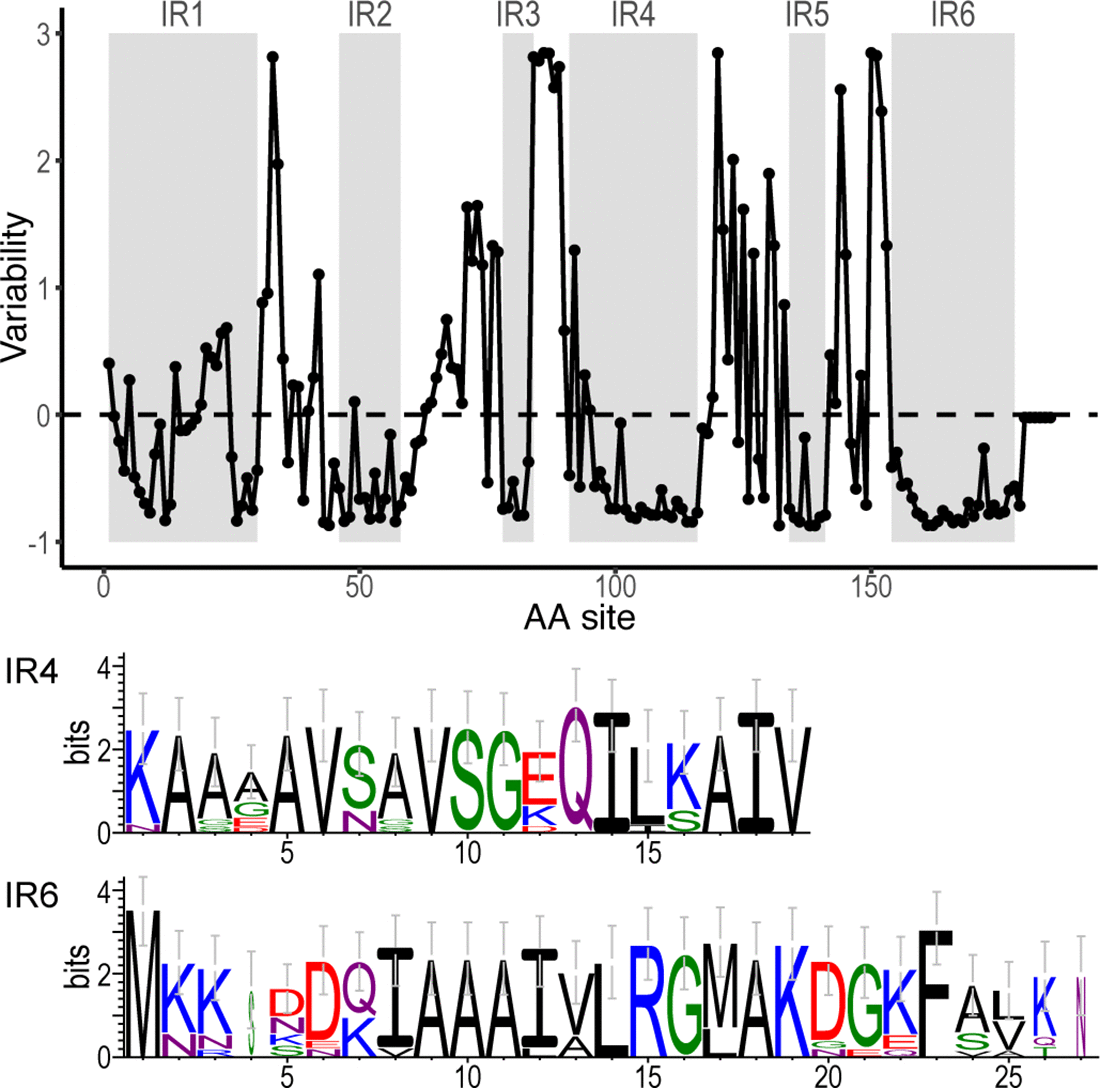
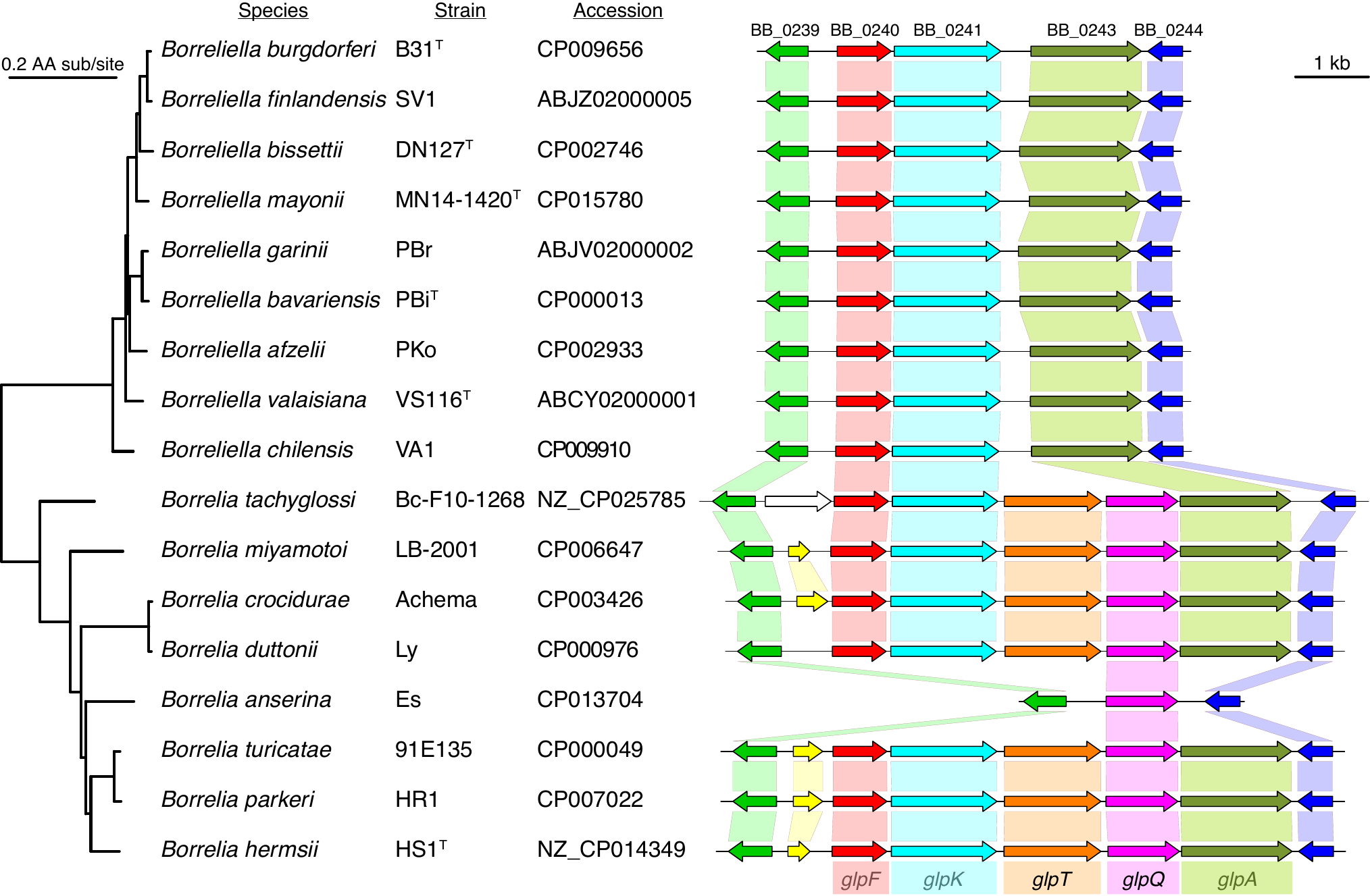
= | File:Ira-fig2.png | link=https://doi.org/10.21775/9781913652616 | Schwartz, Margos, Casjens, Qiu, Eggers (2020). "Multipartite Genome of Lyme Disease ''Borrelia'': Structure, Variation and Prophages". '''''Current Issues in Molecular Biology'''''. 42:409-454. | ||
= | File:Fig6-1-Barbour.png | link=https://doi.org/10.1002/9781118960608.gbm01525 | Barbour & Qiu (2019). ''Borreliella''. In '''''Bergey's Manual of Systematics of Archaea and Bacteria'''''. John Wiley & Sons, Inc., in association with Bergey's Manual Trust. | ||
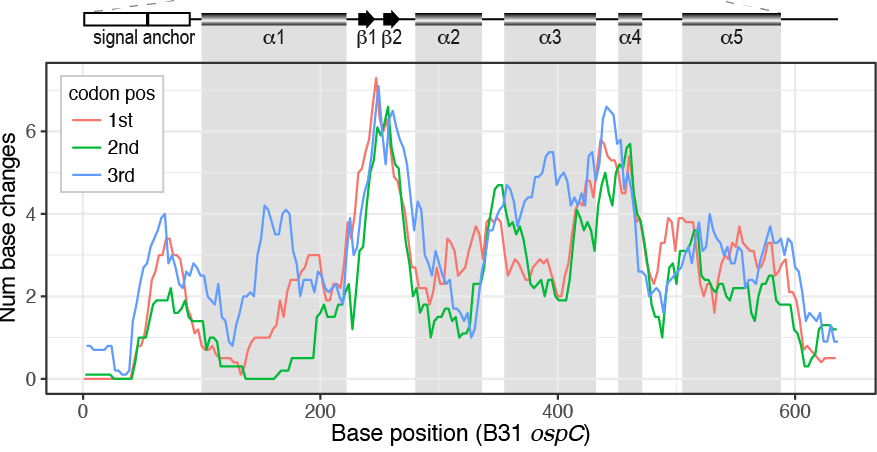
File:Zjm9990961420002.jpeg | link=https://doi.org/10.1128/jcm.00940-18 | Di L, Wan Z, Akther S, Ying CX, Larracuente A, Li L, Di C, Nunez R, Cucura DM, Goddard NL, Krampis K, Qiu WG. (2018). Genotyping and Quantifying Lyme Pathogen Strains by Deep Sequencing of the Outer Surface Protein C (''ospC'') Locus. '''Journal of Clinical Microbiology'''. 56(11):e00940-18. | |||
</gallery> | |||
===Evolution & Learning Algorithms=== | |||
=== | <gallery mode="packed" heights="200px" perrow="3" style="text-align:left"> | ||
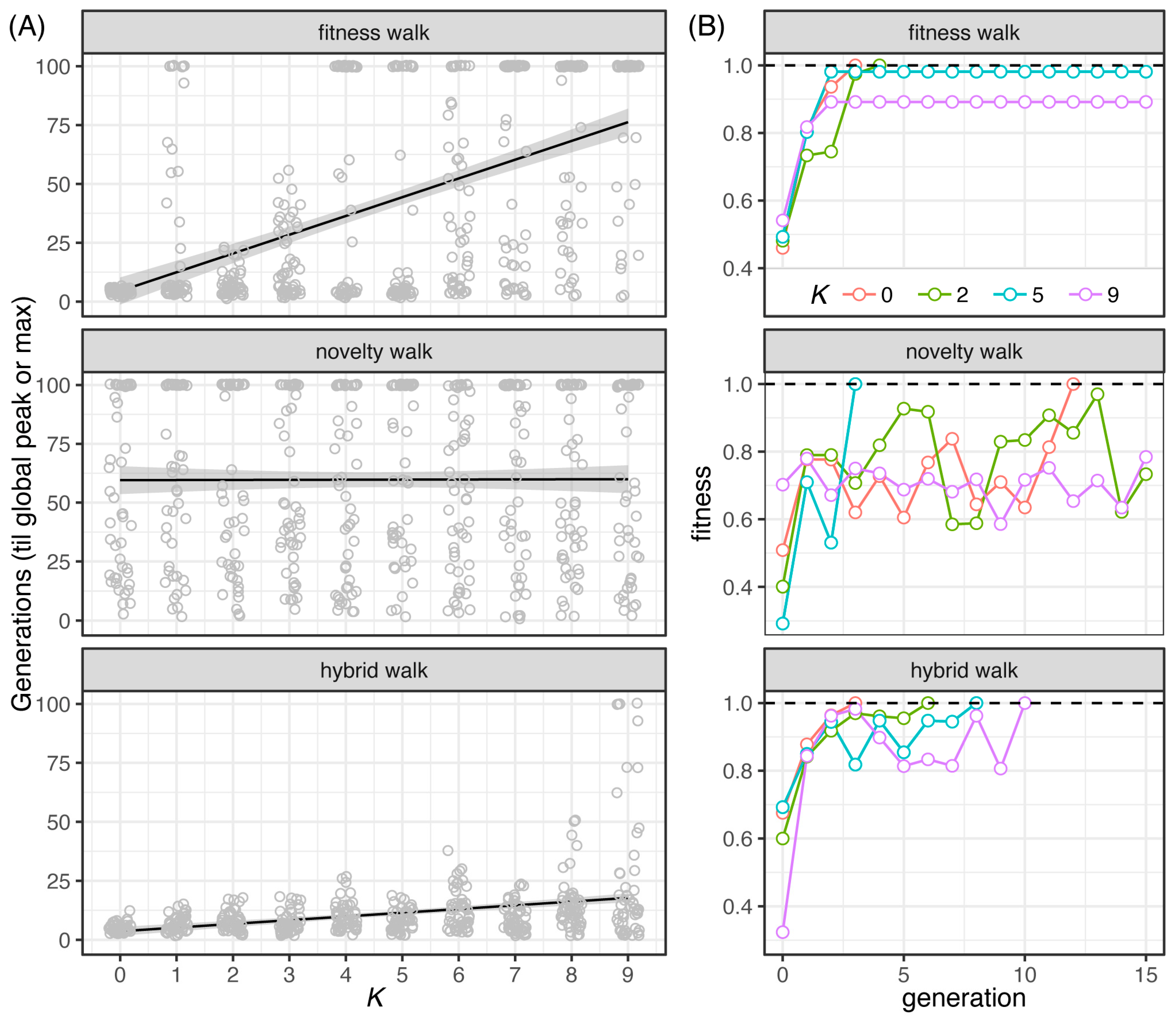
File:Pathogens-12-00388-g003.png | link=https://doi.org/10.3390/pathogens12030388 | Ely, Koh, Ho, Hassan, Pham, Qiu (2023). Novelty Search Promotes Antigenic Diversity in Microbial Pathogens. '''''Pathogens''''' 12:388. | |||
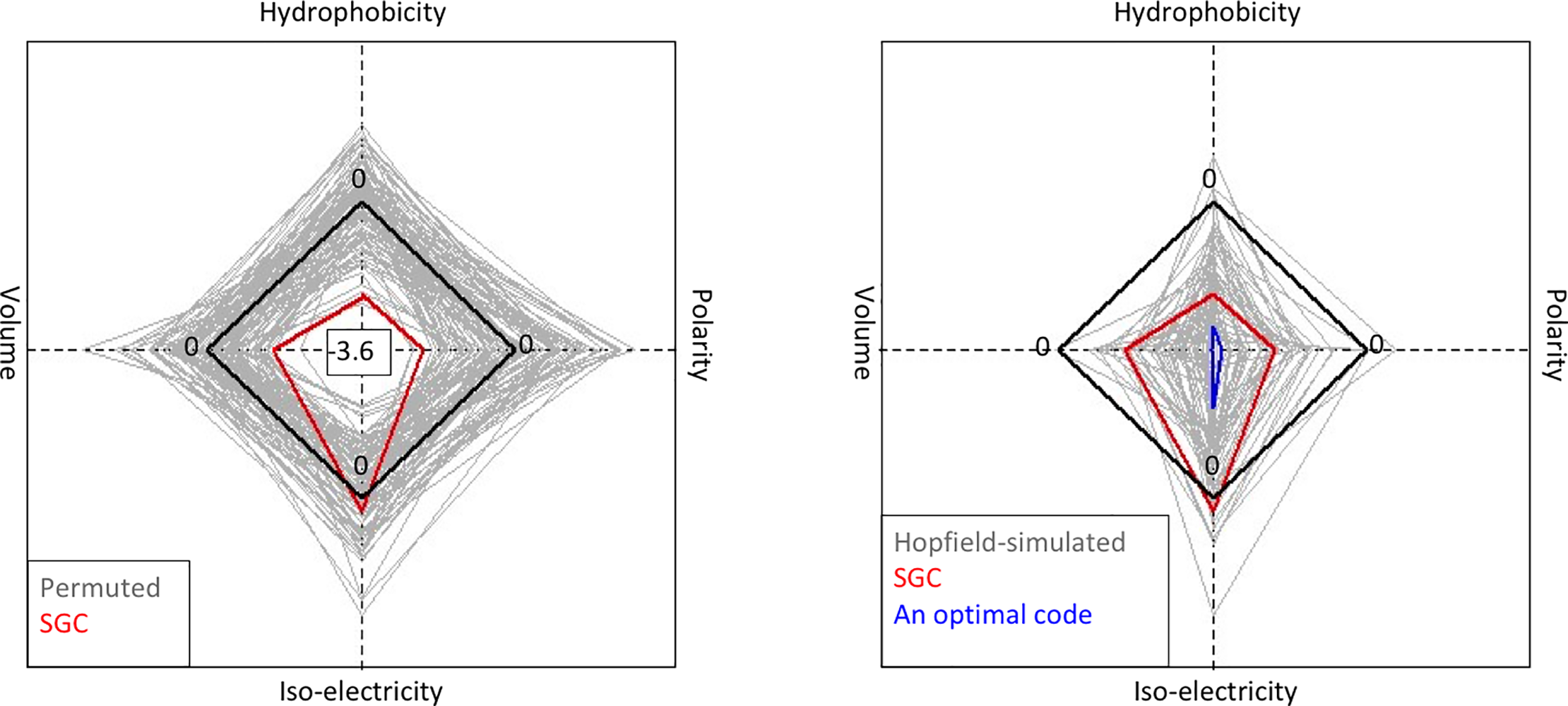
File:Pone.0224552.g005.png | link=https://doi.org/10.1371/journal.pone.0224552 | Attie, Sulkow, Di, Qiu (2019). Genetic codes optimized as a traveling salesman problem. '''''PLoS ONE''''' 14(10): e0224552. | |||
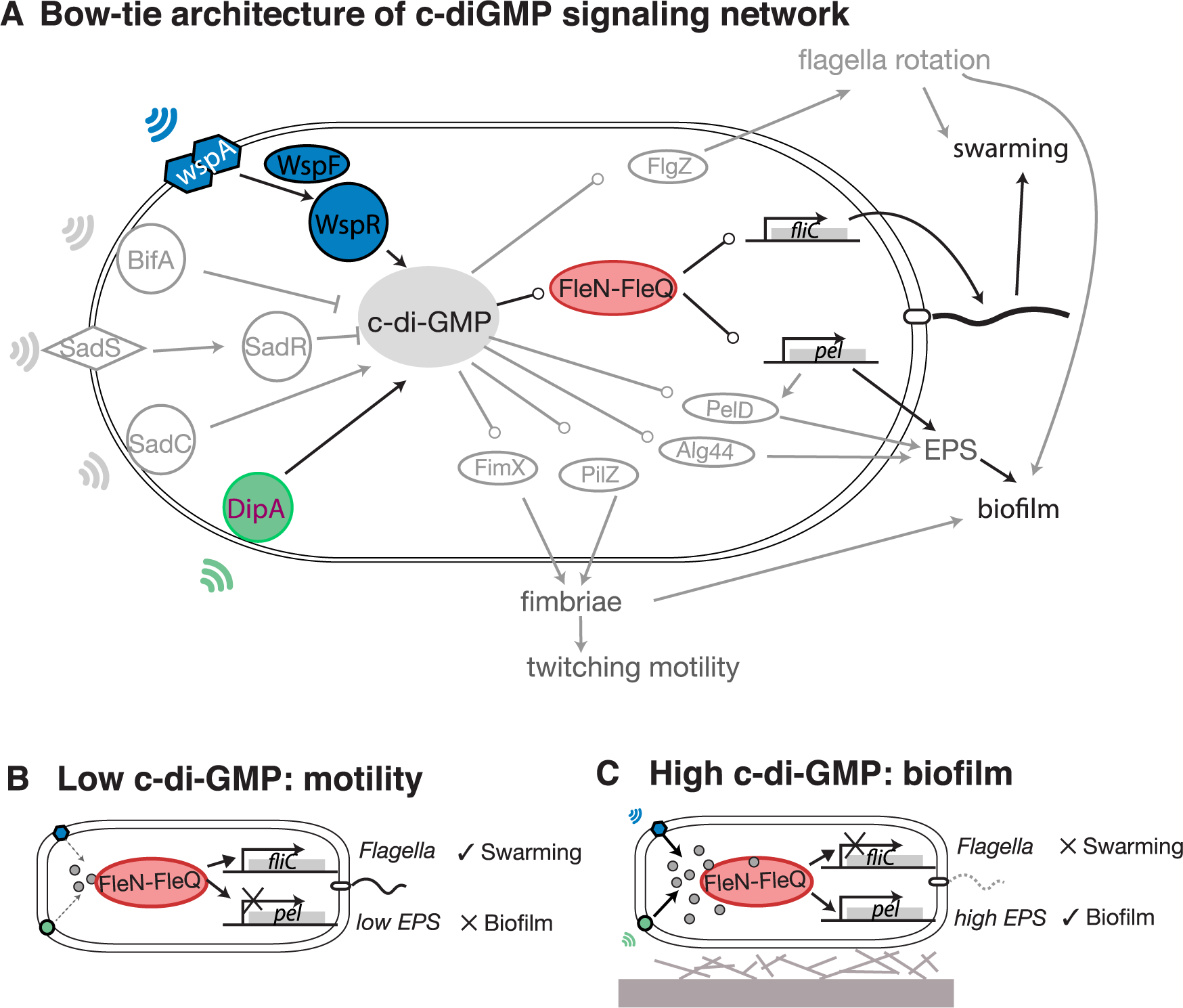
File:Pcbi.1005677.g001.png | link=https://doi.org/10.1371/journal.pcbi.1005677 | Yan, Deforet, Boyle, Rahman, Liang, Okegbe, Dietrich, Qiu, Xavier (2017). Bow-tie signaling in c-di-GMP: Machine learning in a simple biochemical network. '''''PLoS Comput Biol''''' 13(8): e1005677. | |||
</gallery> | |||
* | ===Informatics Tool Development=== | ||
* | <gallery mode="packed" heights="200px" style="text-align:left"> | ||
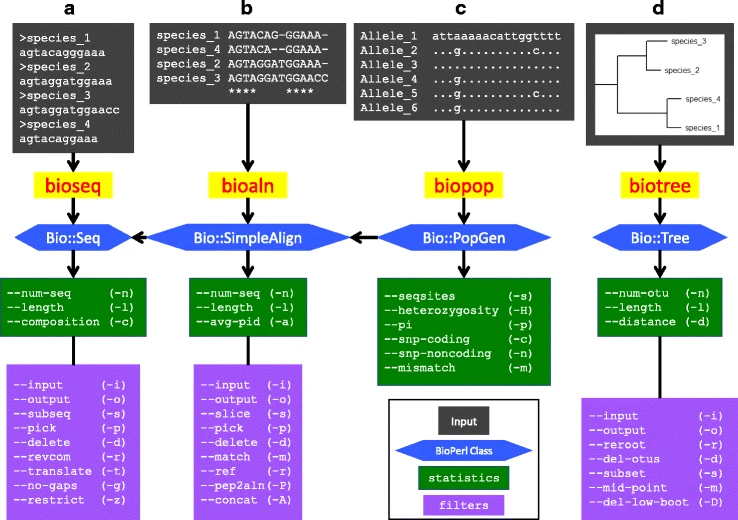
* | File: 12859 2018 2074 Fig1 HTML.png | link=https://pubmed.ncbi.nlm.nih.gov/29499649 | Hernandez, Bernstein, Pagan, Vargas, McCaig, Ramrattan, Akther, Larracuente, Di, Vieira, Qiu. (2018). BbWrapper: BioPerl-based sequence and tree utilities for rapid prototyping of bioinformatics pipelines. '''''BMC Bioinformatics''''' 19(1):76. | ||
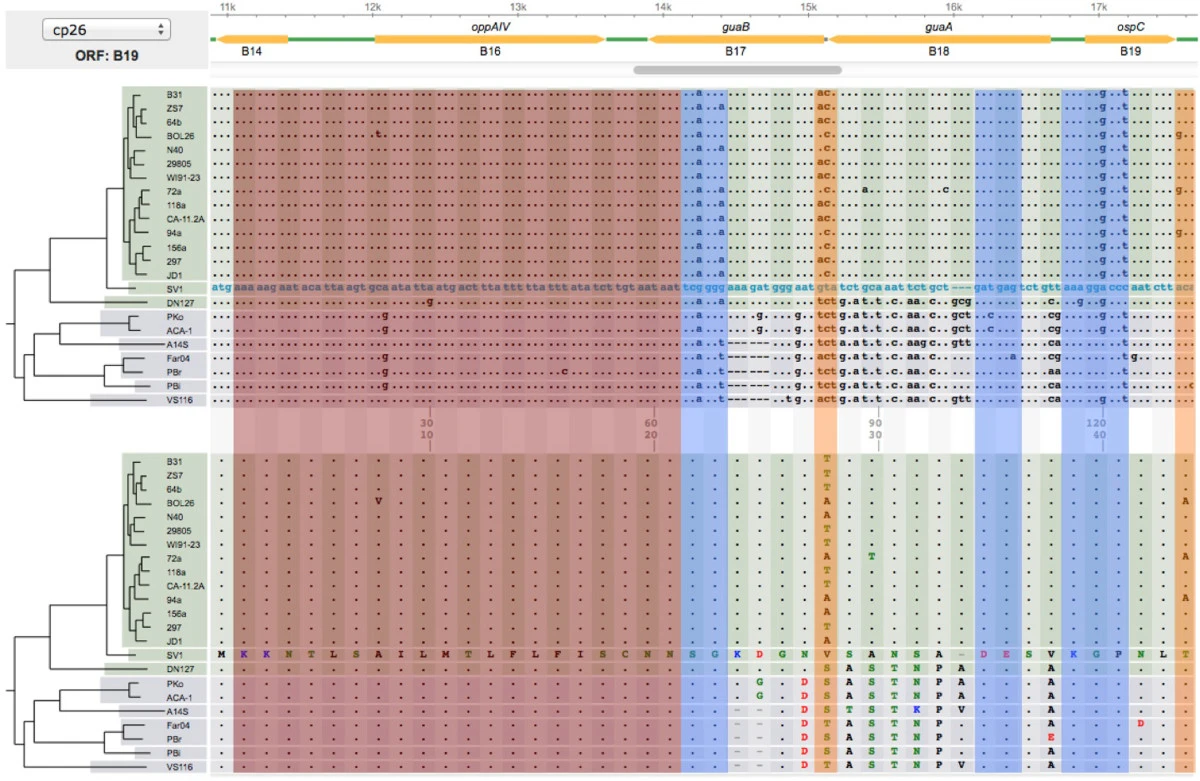
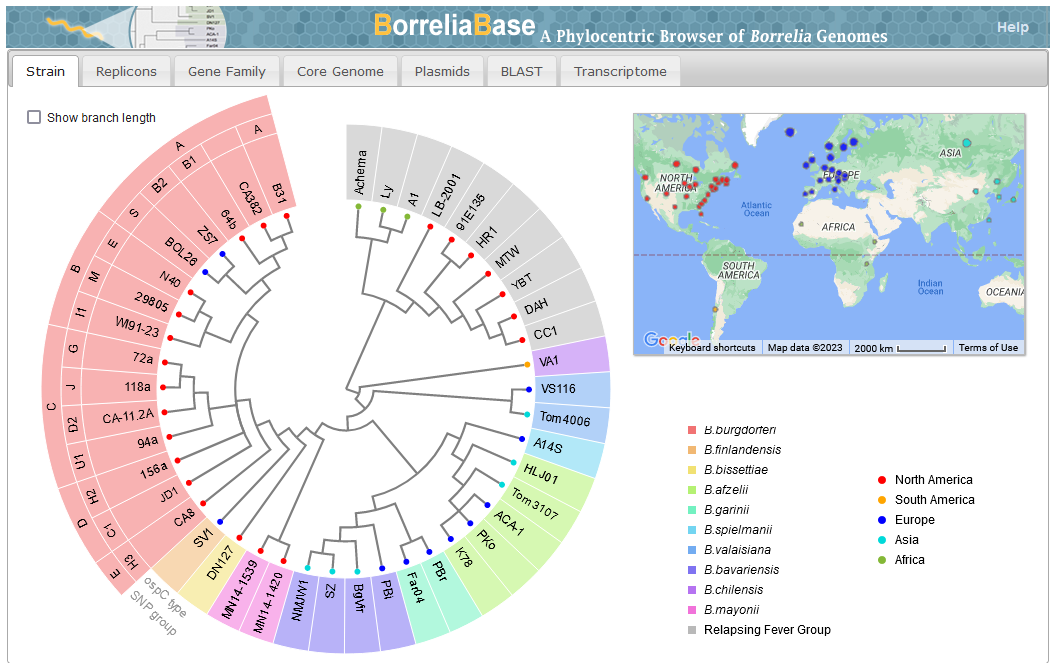
* | File: 12859 2014 Article 6488 Fig3 HTML.png | link=https://pubmed.ncbi.nlm.nih.gov/24994456 | Di, Pagan, Packer, Martin, Akther, Ramrattan, Mongodin, Fraser, Schutzer, Luft, Casjens and Qiu. (2014). BorreliaBase: a phylogeny-centered browser of ''Borrelia'' genomes. '''''BMC Bioinformatics''''' 15:233. | ||
* | </gallery> | ||
* | |||
* | * [http://scholar.google.com/citations?hl=en&user=Ds6u39QAAAAJ) Full list by Google Scholar] | ||
* [http://www.ncbi.nlm.nih.gov/sites/myncbi/weigang.qiu.1/bibliography/42770924/public/ Full list by NCBI Bibliography] | |||
last update: March 20, 2023 | |||
==Lab members and trainees== | |||
{| class="wikitable" | |||
|- | |||
! Year/Period !! Doctoral members & trainees !! Other members & trainees | |||
|- | |||
| Current Academic Year | |||
(Fall 2022, Spring 2023 & Summer 2023) | |||
* Spring | || | ||
* | * Brandon Ely: CUNY Grad Center, Biology/MCD doctoral program | ||
* | * Dr Yozen Hernandez: System administrator (part-time), Ph.D. from Boston University | ||
** [[ | || | ||
* | * Tasmina Hassan: Hunter Bio/CS | ||
* | * Mathew DiCicco: Hunter Math/CS | ||
* | * Tara Doma Lama: Hunter Bio/Bioinformatics | ||
* | * Hagar Abuzaid: Hunter Bio | ||
* | |- | ||
** [[ | | Alumni (Since Fall 2002) | ||
** [ | || | ||
* Li Li (Lily, 2023): CUNY Grad Center, Biology/EEB doctoral program | |||
* Dr Lia Di: Ph.D. Postdoctoral Research Associate, from Nanjing Agricultural University & Wisconsin Blood Institute | |||
* Bioinformatics | * Dr Saymon Akther (2022): CUNY Grad Center, Biology/EEB | ||
* Dr Rayees Rahman: Hunter Bio/Bioinformatics, Ph.D. from Mt Sinai Medical School | |||
---- | * Dr Che Martin (2013): CUNY Grad Center, Biology/MCD | ||
* Dr James Haven (2011): CUNY Grad Center, Biology/MCD | |||
* Dr Tika Sukarna (2009): CUNY Grad Center, Biology/MCD | |||
* Dr Juan Coronado (2008): CUNY Grad Center, Biology/MCD (Dr Peter Lipke) | |||
* Dr William McCaig: CUNY BA, Ph.D. from Stony Brook University | |||
* Dr Vincent Xue: CUNY CS/Bioinformatics, Ph.D. from MIT | |||
* Dr Fubin Li: CUNY Grad Center, Biology/MCD (Dr Laurel Eckhardt) | |||
* Dr Oliver Attie: Postdoctoral Research Associate, Ph.D. from NYU | |||
|| | |||
(published coauthors)<br> | |||
* Brian Sulkow: CUNY Grad Center, Mathematics | |||
* Winston Koh: Hunter Bio/CS | |||
* Eamen Ho: Hunter Bio/Bioinformatics | |||
* Ahn Pham: Hunter Bio/Bioinformatics | |||
* Chris Panlasigui: Hunter Bio/Bioinformatics | |||
* Amanda Amanda Larracuente: CUNY Grad Center, Biology/MCD | |||
* Pedro Pagan: Hunter Bio/Bioinformatics | |||
* Edgaras Bezrucenkovas: Hunter Chem/Bioinformatics | |||
* Girish Ramrattan: Hunter Bio/Bioinformatics | |||
* Levy Vargas: Hunter Bio/Bioinformatics | |||
* Chong Di: Hunter Geography | |||
* Roy Nunez: Hunter Bio | |||
* Mei Wu: CUNY City Tech | |||
* Desiree Pante: Hunter Bio | |||
* Saimtun Shipa: Hunter Stat (MA) | |||
* Bing Wu: Hunter Bio/Biotechnology | |||
* Svidatoslav Kendall (Slav): Hunter Biology | |||
* Philip Romov: Hunter CS | |||
|} | |||
==Web Apps== | |||
===Apps with Collaborators (or from published papers)=== | |||
<gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | |||
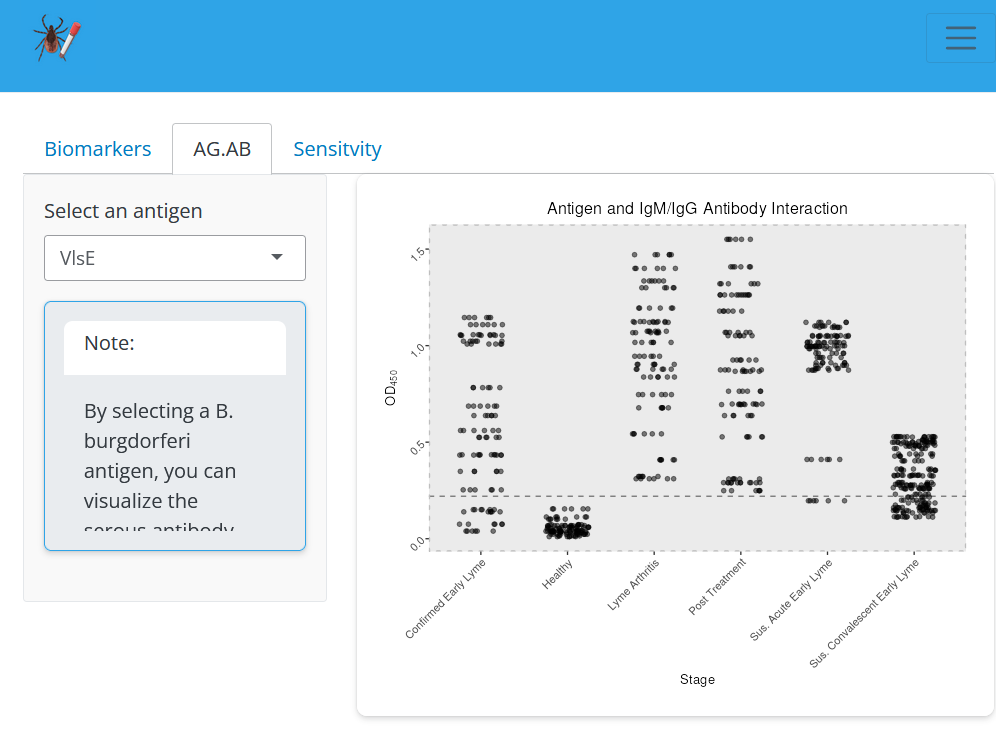
File:Screenshot_2024-06-19_205621.png | link=https://cov.genometracker.org/finalelisa-app/ | Borrelia diagnostic antigens (App developed by Liann Aris-Henry; Data from [https://journals.asm.org/doi/10.1128/jcm.01142-19 Arumugam et al (2019)]) | |||
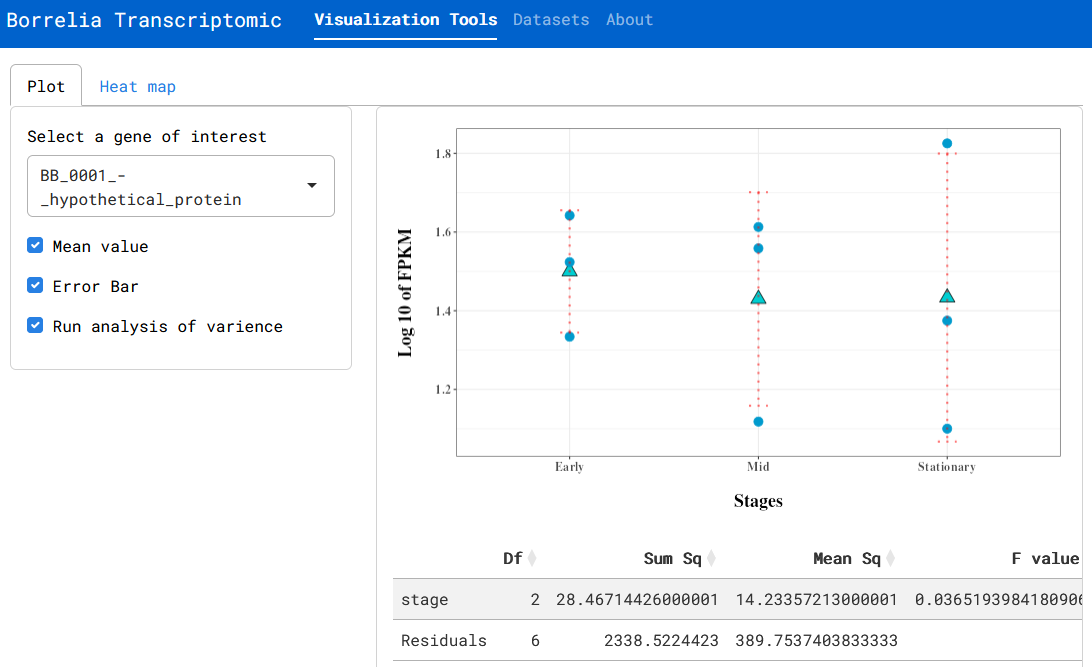
File:Screenshot_2024-06-18_144931.png | link=https://cov.genometracker.org/borrelia-app/ | Borrelia growth transcriptome (App developed by Laziz Asamov; Data from [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0164165 Arnold et al (2016)]) | |||
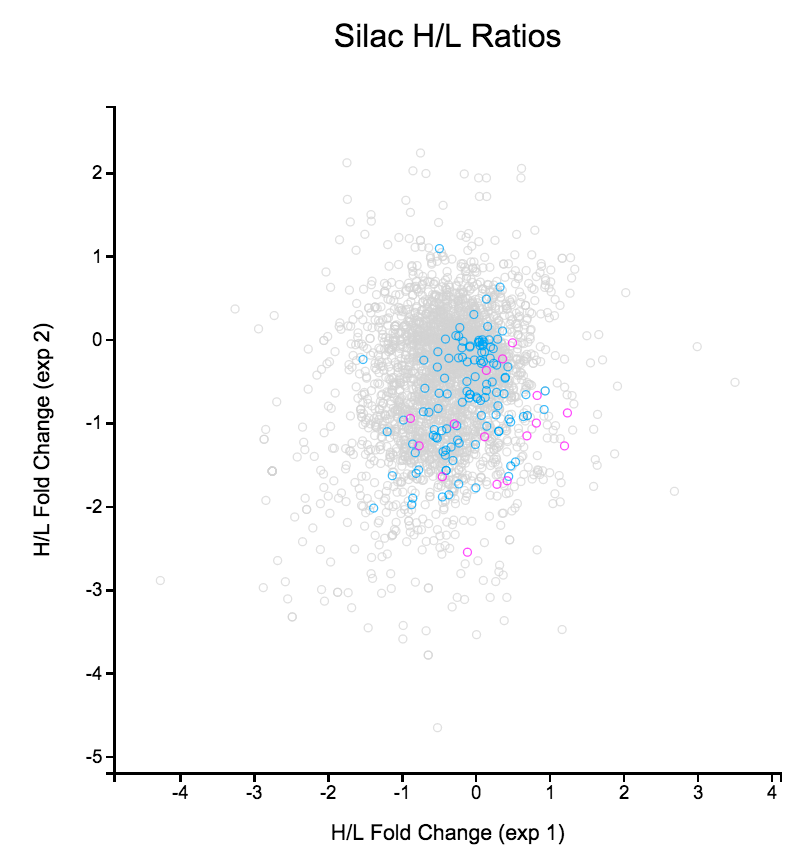
File: Silac.png | link=http://borreliabase.org/~wgqiu/Polotskaia_etal_2014/supp-fig-s1/ | Data & [http://borreliabase.org/~wgqiu/Polotskaia_etal_2014/supp-table-s1/ GSEA] associated with Polotskaia_etal_2014 (with Bargonetti Lab @Hunter) | |||
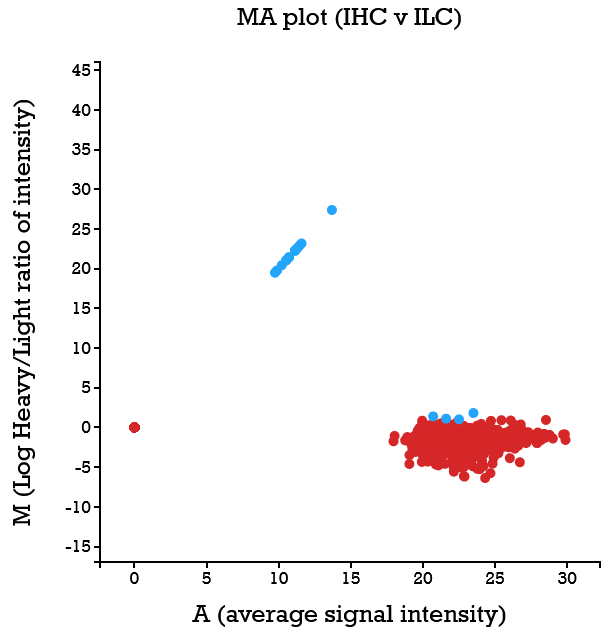
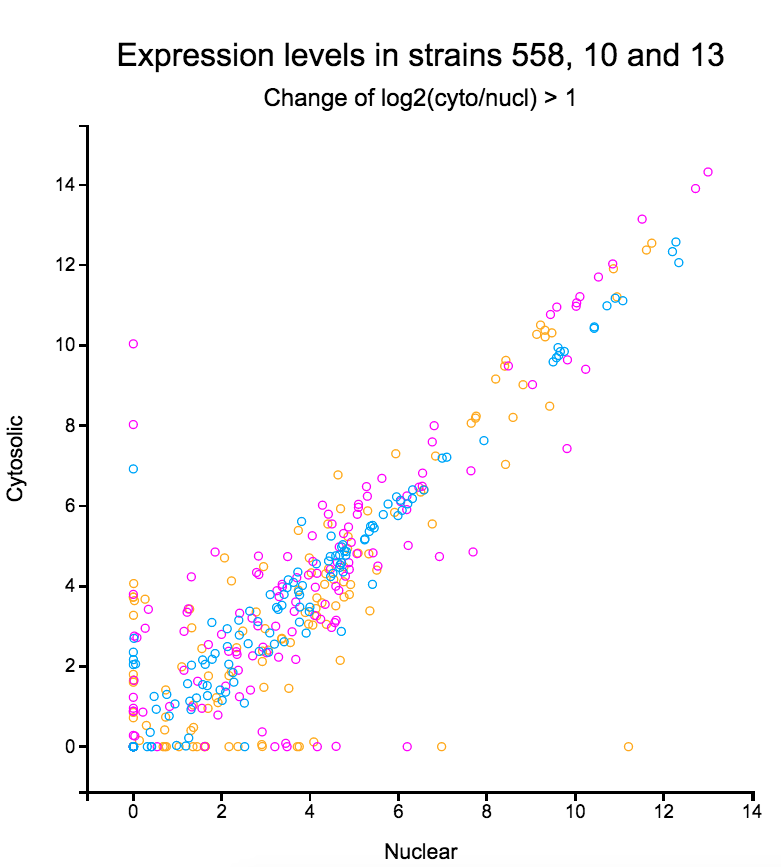
File:Screenshot_2023-03-24_122016.png | link=http://borreliabase.org/~wgqiu/clickme-khalikuz/temp-Points.html | Phosphoproteome MDM2KD data set (with Bargonetti Lab @Hunter) | |||
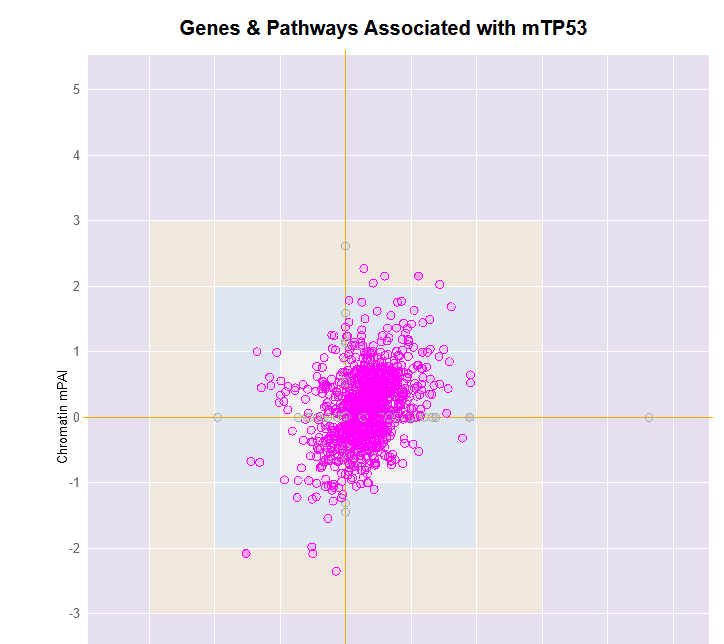
File:Screenshot_2023-03-24_122312.png | link=http://borreliabase.org/~wgqiu/mpai-v3/ | Genes & Pathways Associated with mTP53 (with Bargonetti Lab @Hunter) | |||
File:Spombe.png | link=Spombe | S. pombe transcriptomes (with Zhong Lab @Hunter) | |||
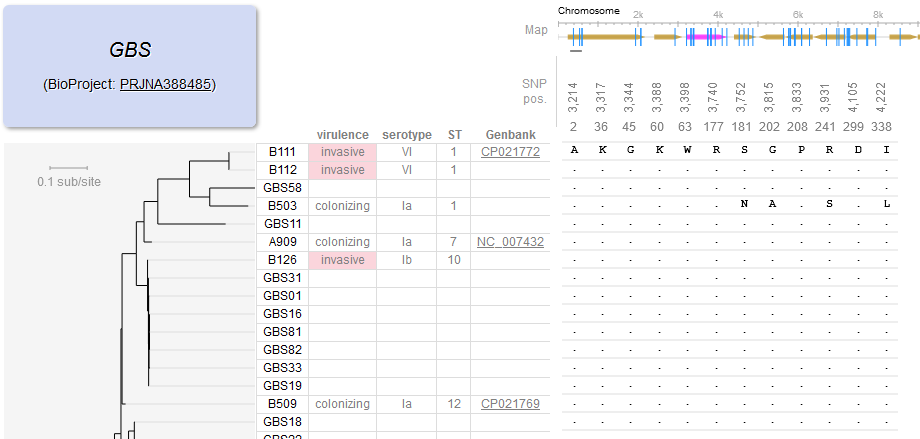
File:Screenshot 2023-03-24 122223.png | link=http://borreliabase.org/~wgqiu/gbs-browser-v3/ | Group B Streptococcus (GBS) genome browser (with Wu Lab @Shenzhen) | |||
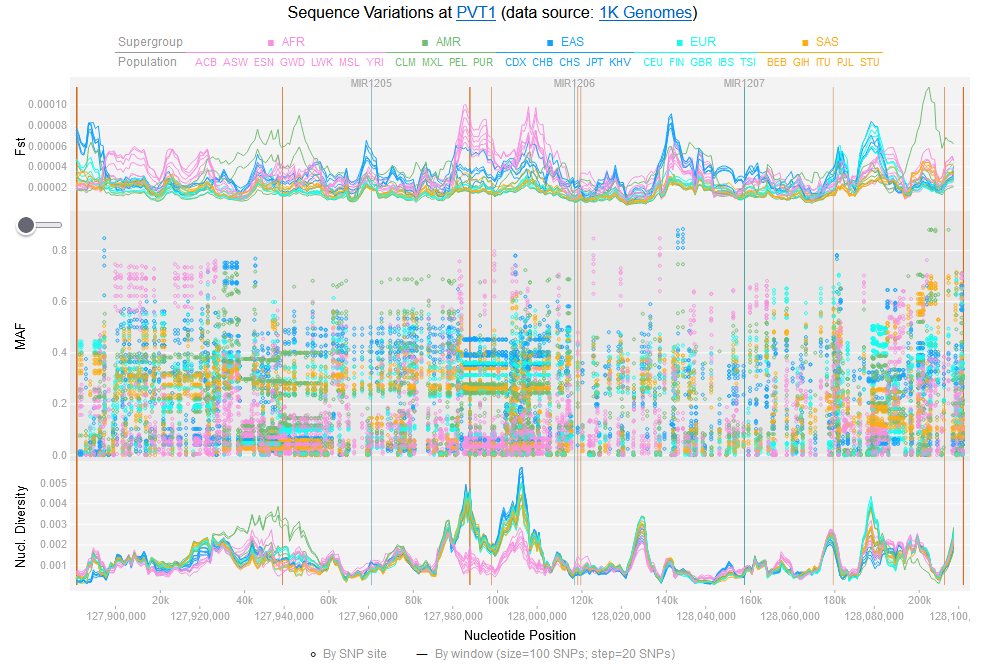
File:Screenshot 2023-03-24 122245.png | link=http://borreliabase.org/~wgqiu/oneKGenome/ | 1K genome (with Ogunwobi Lab @Hunter) | |||
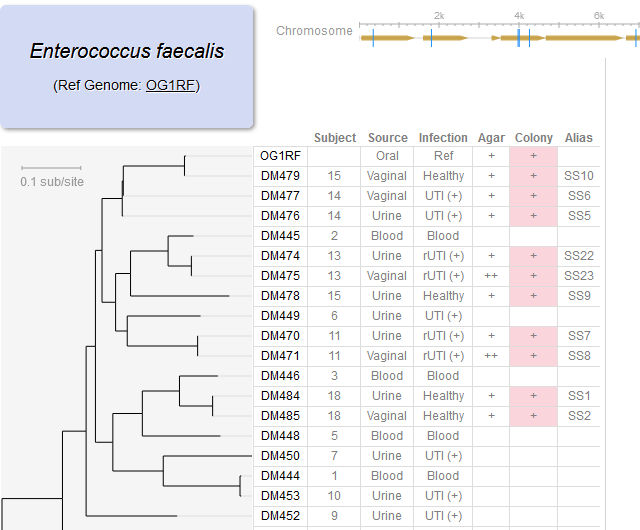
File:Screenshot 2023-03-24 122450.png | link=http://borreliabase.org/~wgqiu/E_faecalis/ | E faecalis genome browser (with Morales Lab @WCMC) | |||
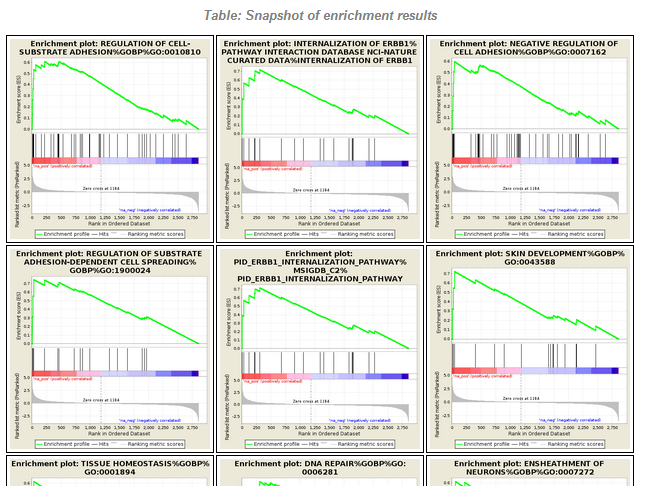
File:Screenshot 2023-03-24 122145.png | link=http://borreliabase.org/~wgqiu/carmen-proteomics/ | Proteomics GSEA results (with Melendez Lab @Hunter) | |||
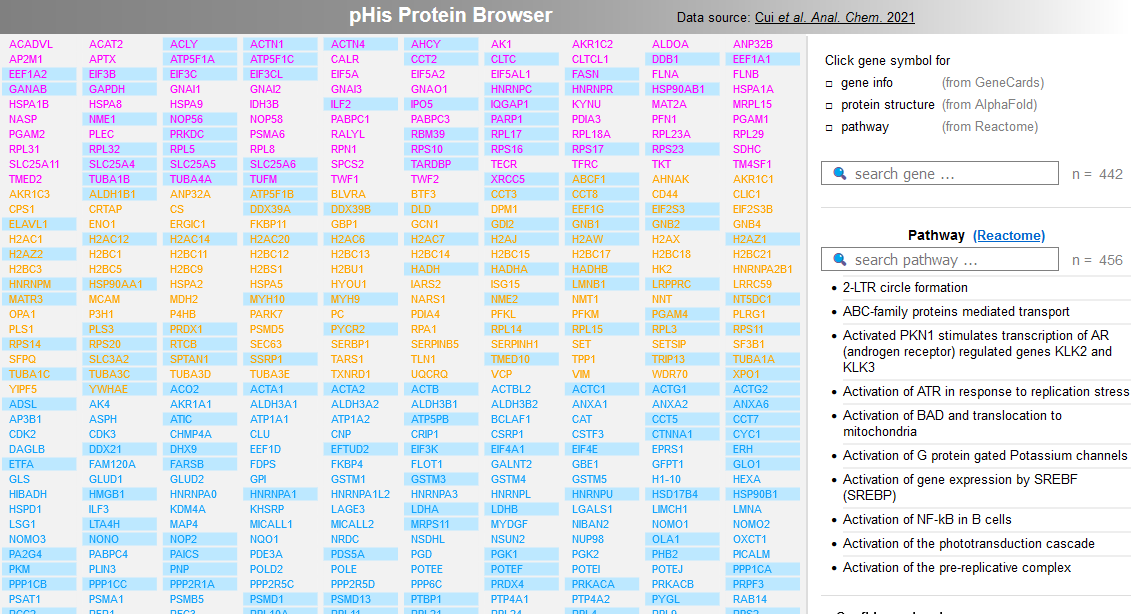
File:Screenshot 2023-03-24 122051.png | link=http://borreliabase.org/~wgqiu/Tcell-pHis-ACS/ | pHis Protein Browser (with Skolnik lab @NYU) | |||
</gallery> | |||
===Qiu Lab Apps=== | |||
<gallery mode="packed" heights="200px" perrow="3" style="text-align:left"> | |||
File:Screenshot 2023-03-24 122558.png | link=http://borreliabase.org | Lyme genome browser | |||
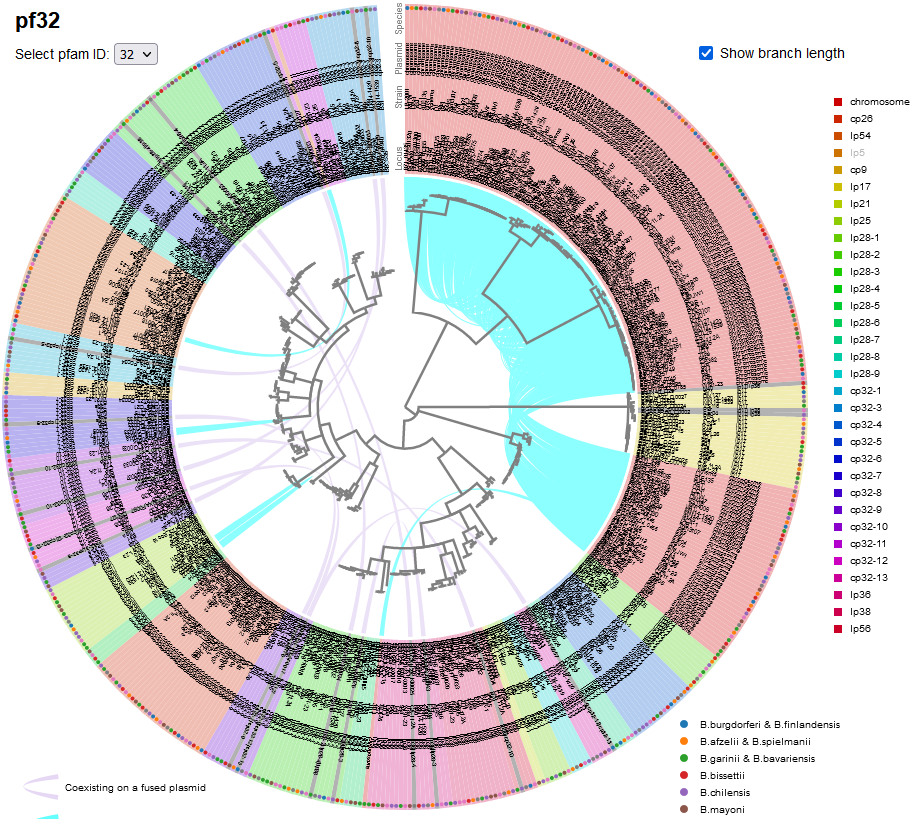
File:Screenshot 2023-03-24 133320.png | link=http://borreliabase.org/~wgqiu/pf-trees/ | Lyme pathogen plasmid partitioning gene trees | |||
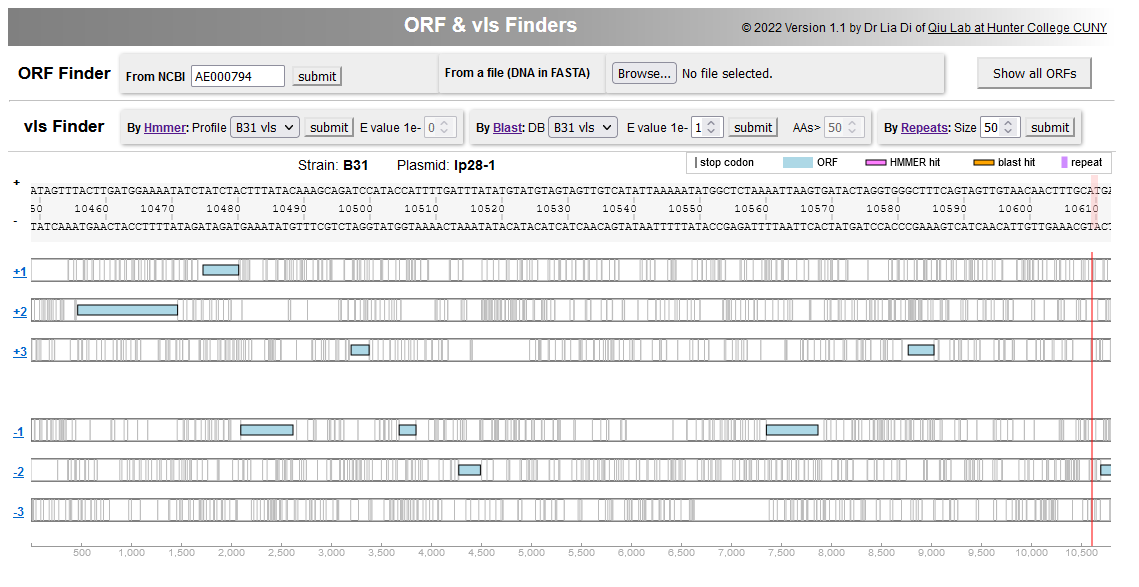
File:Screenshot 2023-03-24 133636.png | link=http://borreliabase.org/vls-finder/ | vls Finder in Lyme pathogen genomes | |||
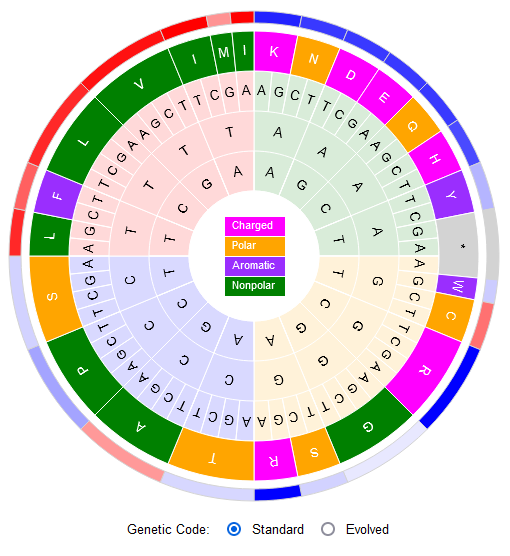
File:Screenshot 2023-03-24 122359.png | link=http://borreliabase.org/~wgqiu/code-wheel | Codon Wheel | |||
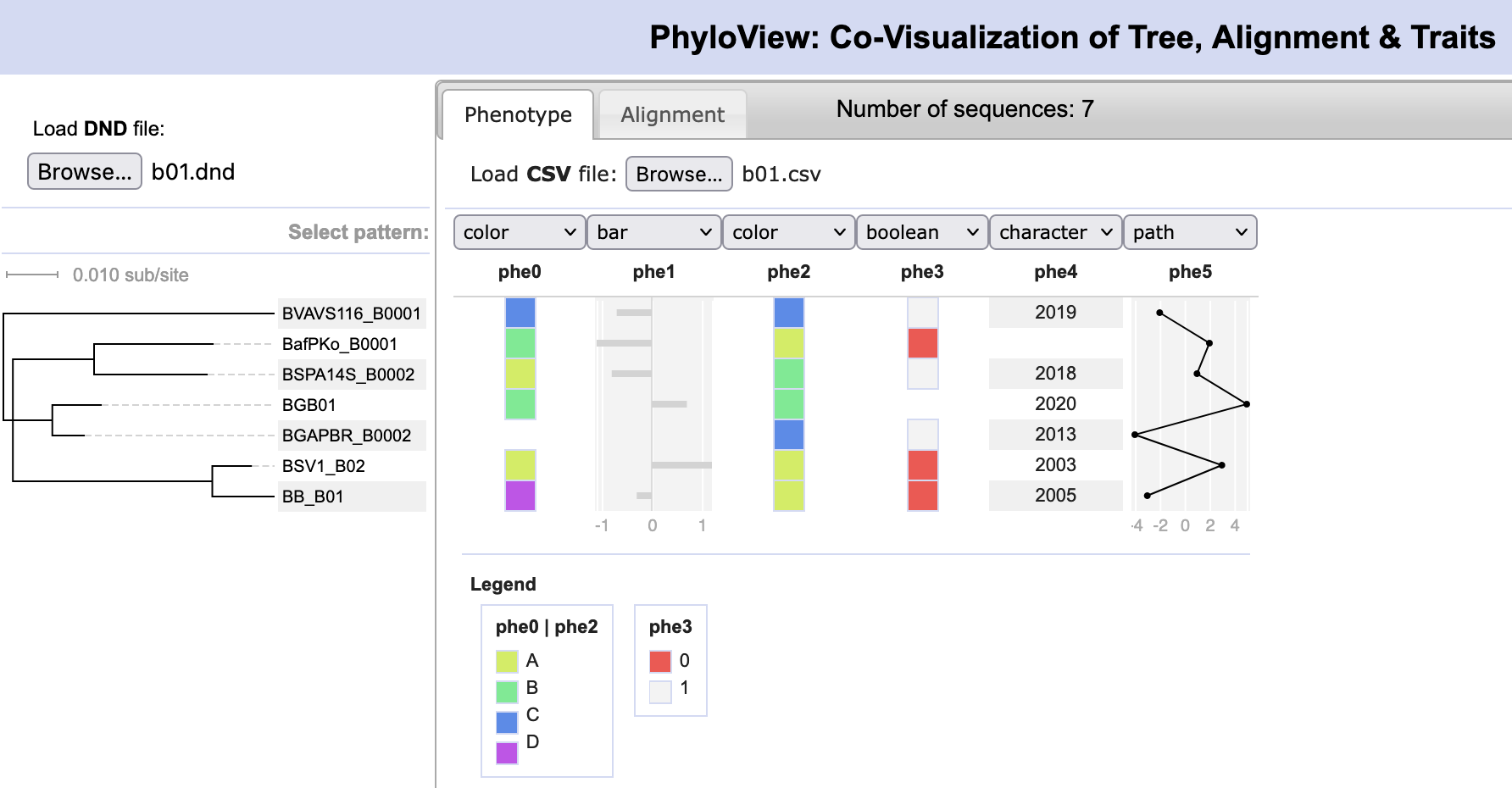
File:PhyloView.png | link=http://borreliabase.org/~wgqiu/PhyloView | Co-visualization of a tree with an alignment and character matrix | |||
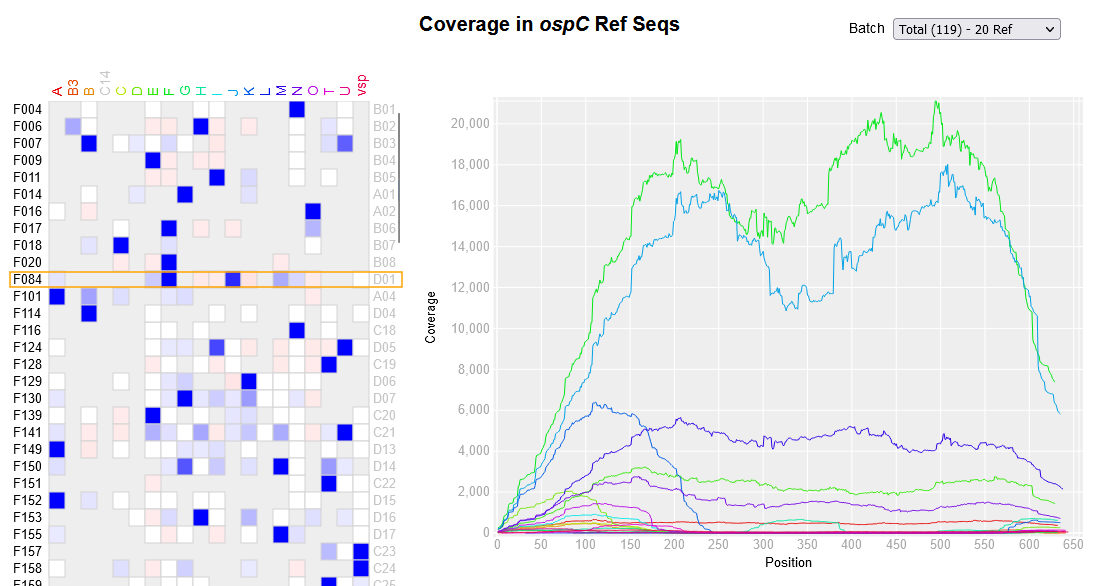
File:Screenshot 2023-03-24 122521.png | link=http://borreliabase.org/~wgqiu/ospC-sequencing | OspC amplicon sequencing from ticks | |||
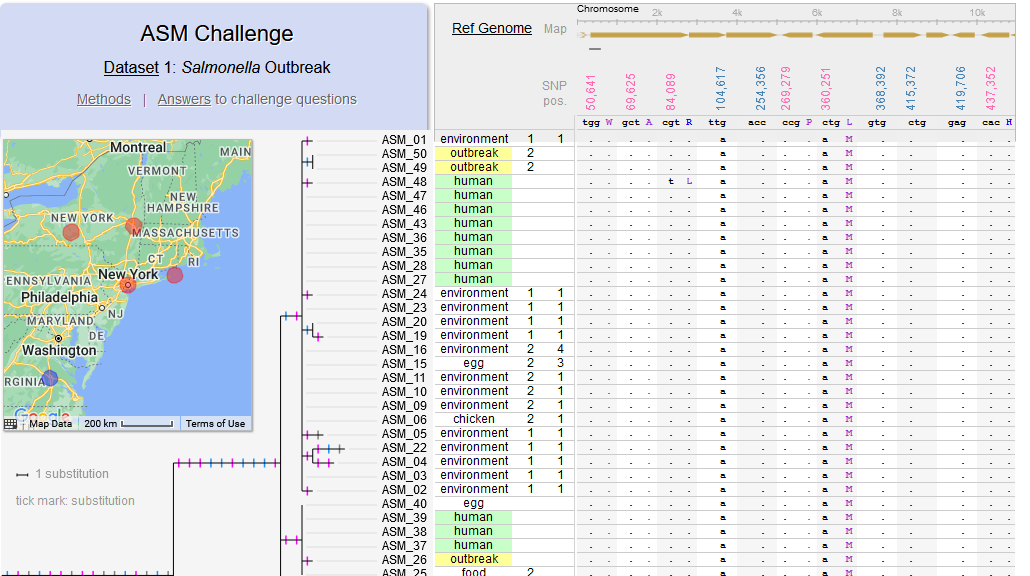
File:Screenshot 2023-03-24 122340.png | link=http://borreliabase.org/~wgqiu/asm-challenge | Genomic epidemiology of a Salmonella outbreak (ASM Challenge) | |||
</gallery> | |||
==Curricular Development & Bioinformatics/QuBi Advising== | |||
* QuBi advisors: Weigang Qiu, Ntino Krampis, Rabindra Mandal (Biology); Saad Mneimeih, Lei Xie (CS); Akira Kawamura (Chem); Dana Sylvan (Math & Stats) | |||
** Permission for non-Biology majors to take BIOL203 & BIOL425, every Spring | |||
** Collect names, major, and IDs to send to course coordinator to grant permission. Waive BIOL10200 pre-reqs for taking BIOL203. | |||
* Curricular resources: | |||
** [http://biology.hunter.cuny.edu/index.php?option=com_content&view=article&id=66&Itemid=73 Biology courses and pre-reqs] | |||
** [https://hunter-undergraduate.catalog.cuny.edu/programs/BIO1-BA Hunter Biology Major 1 (including the Bioinformatics Option)] | |||
** [https://hunter-undergraduate.catalog.cuny.edu/programs/CHEM2-BA Hunter Chemistry Major 2 (including the Bioinformatics Option)] | |||
** [https://hunter-undergraduate.catalog.cuny.edu/programs/COMPSCI-BA Hunter Computer Science (including Bioinformatics Concentration)] | |||
** [https://hunter-undergraduate.catalog.cuny.edu/programs/MATH-BA Hunter Mathematics (including the Quantitative Biology Concentration)] | |||
** [https://hunter-undergraduate.catalog.cuny.edu/programs/STATS-BA Hunter Statistics (including the Quantitative Biology Concentration)] | |||
* QuBi advising: | |||
** Declaration of Bioinformatics concentration: In-person advising to work out the semester-by-semester courses | |||
** Approve on department spreadsheet (or send email to "Samantha Sheppard-Lahiji" and "HTR Bio" <biology@hunter.cuny.edu>) | |||
** Students should take Bioinformatics-specific electives (8 cred; see Hunter Catalog below), '''not general electives''' | |||
*** Examples: Anthrop302 (3 cr); Chem333 (3 cr); BIOL47119 & BIOL47120 (3cr); BIOL48002 (2 cr) | |||
** Students need to take BIOL48002 (2 cr), which counts towards as research credit, to graduate as honors | |||
* General advising: | |||
** ~40 students every semester. Send out emails to students. Go through student courses by Email or by appointment | |||
** Recommend new math courses: '''MATH15200 & STAT21350''' | |||
* Hosting QuBi students in lab | |||
** This is to enhance the informatics and coding skills of our students | |||
** Students should register and get '''BIOL48002''' credits, which counts towards their elective credits & eligibility for honors | |||
** 3-5 students per semester | |||
* Outside research opportunities | |||
** MIT Quantitative Workshop (first week of January, in Boston). Coordination with CS (Saad & Susan Epstein) in Fall | |||
** Simons Foundation/Flatiron Institute Center for Computational Biology (CCB) Internship program. Open House in Spring | |||
==Course/Lecture syllabi== | |||
* [[NYRaMP-Informatics-2025|NYRaMP Workshop (August 2025, by Brandon Ely)]] | |||
* [[Computational Genomics (KIZ, Fall 2024)]] | |||
* [[NYRaMP-Informatics-2024|NYRaMP Workshop (August 2024)]] | |||
* BIOL47120 BioMedical Genomics (Spring 2024). Tutorials: [https://borreliabase.org/~wgqiu/tutorial-markdown.html R Markdown] [https://borreliabase.org/~wgqiu/cluster-analysis.html Cluster analysis] [https://borreliabase.org/~wgqiu/scRNA-analysis.html single-cell RNA-seq] | |||
* BIOL425 Computational Molecular Biology (Spring, 2023). [https://github.com/weigangq/CSB-BIOL425/tree/master/lecture-materials Lecture material on github] | |||
* BIOL714 Cell Biology: [http://borreliabase.org/~wgqiu/r-demo-2024.html R Demo (Spring 2024)] [http://borreliabase.org/~wgqiu/r-demo-2023.html R Demo (Spring 2023)] | |||
* QuBi module: [[QuBi/module/bio203-lab12—2022|BIOL20300 Molecular Genetics, Lab 12 (2023)]] | |||
* QuBi module: [[QuBi/modules/biol203-geno-pheno-association-2022|BIOL20300 Molecular Genetics, Lab 13 (2022)]] | |||
* QuBi module: [[QuBi/modules/biol303|BIOL30300 Cell Biology, Bioinformatics Lab (transcriptome analysis)]] | |||
* [[BigData 2020|Big Data (Summer, 2020)]] | |||
* [[BioMed-R-2020|BIOL47120 Biomedical Genomics II (Spring, 2020)]] | |||
** [http://borreliabase.org/~wgqiu/tutorial-markdown.html Tutorial: R Markdown (Spring 2024)] | |||
** [http://borreliabase.org/~wgqiu/cluster-analysis.html Tutorial: Cluster analysis (Spring 2024)] | |||
** [http://borreliabase.org/~wgqiu/scRNA-analysis.html Tutorial: single-cel transcriptome analysis (Spring 2024)] | |||
* [[Biol425 2020|BIOL425 Computational Molecular Biology (Spring, 2020)]] | |||
* [[Biol375 2019|BIOL37500, Molecular Evolution (Fall, 2019)]] | |||
* [[Southwest-University|Southwest University R course (Summer, 2019)]] | |||
* [[Biol20N02 2017|Analysis of Biological Data (Spring, 2017)]] | |||
* [[Bioinformatics_Workshop_2014|Bioinformatics Workshop (Summer, 2014)]] | |||
==SARS-CoV-2 genome evolution== | |||
<gallery mode="packed" heights="200px" perrow="3" style="text-align:left"> | |||
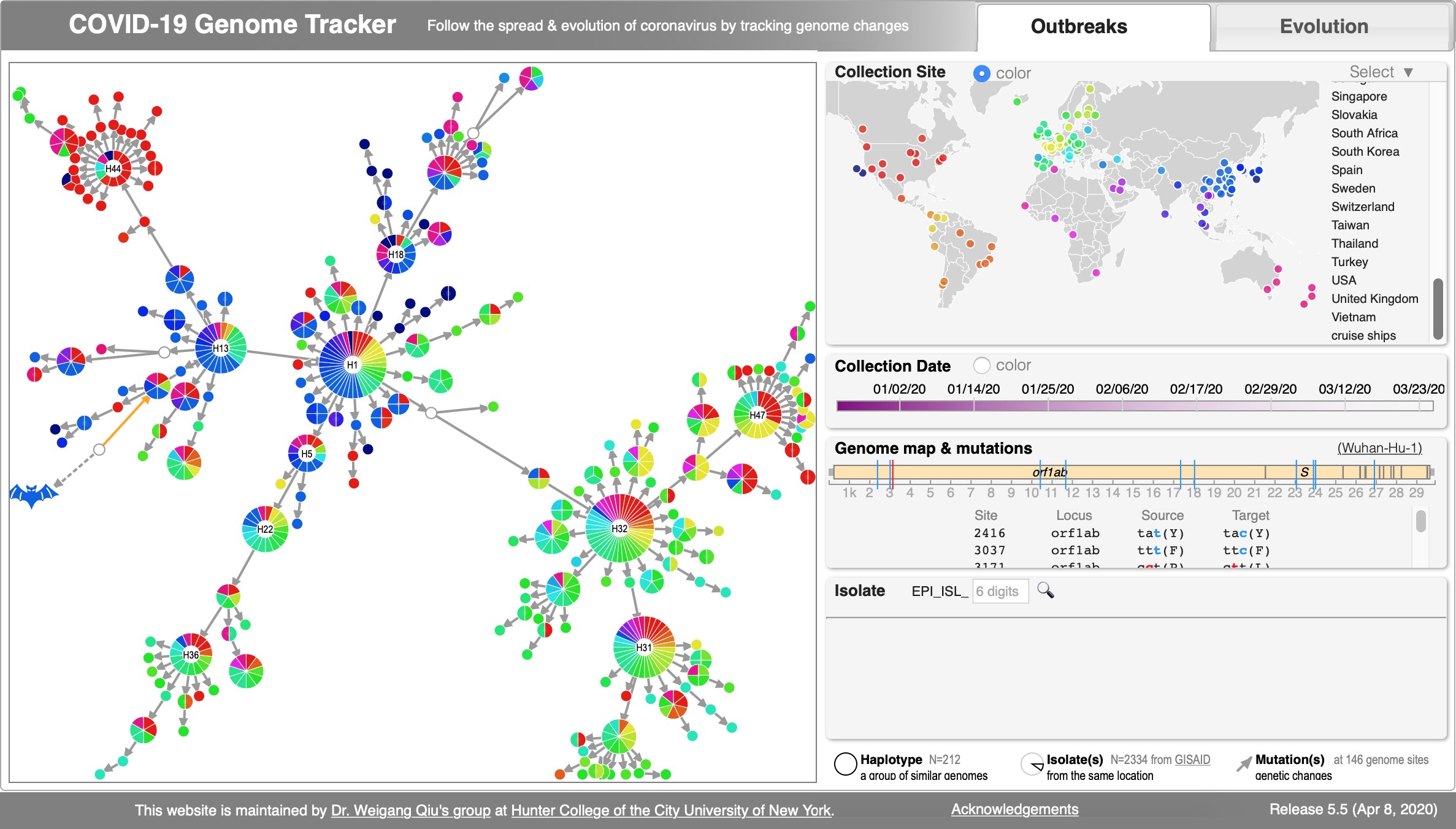
File:Cov-fig1.jpg | Akther, Bezrucenkovas, Sulkow, Panlasigui, Qiu, Di (April, 2020). "CoV Genome Tracker: tracing genomic footprints of Covid-19 pandemic". '''''[https://www.biorxiv.org/content/biorxiv/early/2020/04/14/2020.04.10.036343.full.pdf BioRxiv]'''''; [https://cov.genometracker.org/ Web app: SARS-CoV-2 Genome Tracker]; Github: https://github.com/weigangq/cov-browser | |||
File:Rec-fig2.png | Akther, Li, Martin, Di, Sulkow, Pante, Bezrucenlovas, Luft, Qiu (May, 2020). "Origin, recombination, and missed opprotunities: a genomic perspective of the first 100 days of COVID-19 pandemic". (Unpublished). | |||
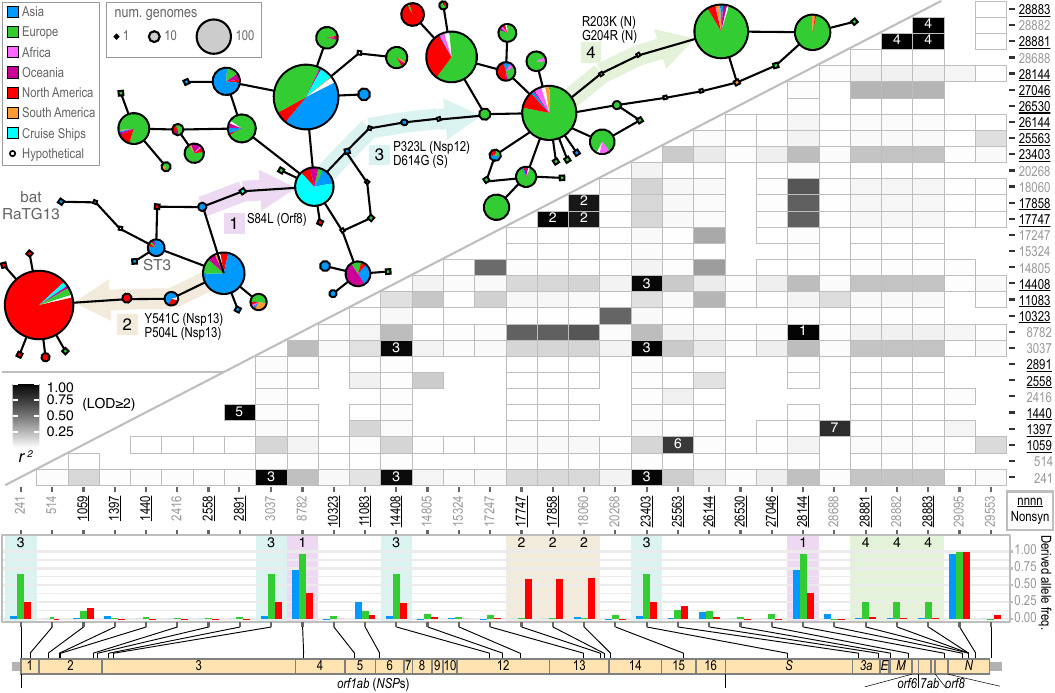
File:Cov-fig3-trace.png | Akther, Bezrucenlovas, Li, Sulkow, Di, Pante, Martin, Luft, Qiu (Sep, 2021). "Following the Trail of One Million Genomes: Footprints of SARS-CoV-2 Adaptation to Humans". '''''[https://www.biorxiv.org/content/biorxiv/early/2021/05/10/2021.05.07.443114.full.pdf BioRxiv link]''''': . Github: https://github.com/weigangq/cov-db | |||
</gallery> | |||
== | ==Lab Resources & Protocols== | ||
* | * [https://runestone.academy/ns/books/published/thinkcspy/index.html How to think like a computer scientist: An interactive Python programming book] | ||
* | * Nanopore sequencing protocols | ||
* | ** DNA barcoding: https://nanopore4edu.org/latest/annotated_experiments/dna_barcoding/ | ||
* | ** Yeast genomes: https://nanoporetech.com/document/extraction-method/yeast-dna | ||
* | * [[Monte Carlo Club]] | ||
* [[NY-RaMP Mentoring]] | |||
* Borreliella genome sequencing consortium: Weekly meetings (Tu @11): Since Jan 2023 | |||
* Borreliella diagnostic antigens (Fall 2023-Fall 2027): | |||
** Zoom call (Jan 23, 2024) | |||
** Next meeting: March 23, 2024 | |||
* Qiu lab network [[First Time Guide|first-time user guide]] | |||
* Qiu lab Github repositories: https://github.com/weigangq/?tab=repositories | |||
* [[Mini-Tutorals|Mini-Protocols]] (frequently used computer codes and pipelines) | |||
* Python tutorial: https://wiki.genometracker.org/~weigang/Intro_to_Python.html | |||
* [[Tick protocol|ick handling protocols]] | |||
* [[A Primer on the Cluster System at Hunter|Hunter HPC Usage]] | |||
* [https://r4ds.hadley.nz/ R for Data Science (2e)], (2024) by Wickham, Grolemund & Çetinkaya-Rundel ([https://bookdown.org/ Bookdown version]) | |||
* Borrelia Genome Consortium: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA431102/ | |||
* Canadian Bbsl genome assemblies: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1130942 | |||
* Nanopore sequencing resources: | |||
** eBook: https://store.nanoporetech.com/us/minion.html | |||
== | == Wiki Help == | ||
* [ | * [https://www.mediawiki.org/wiki/Special:MyLanguage/Manual:Configuration_settings Configuration settings list] | ||
* | * [https://www.mediawiki.org/wiki/Special:MyLanguage/Manual:FAQ MediaWiki FAQ] | ||
* | * [https://lists.wikimedia.org/postorius/lists/mediawiki-announce.lists.wikimedia.org/ MediaWiki release mailing list] | ||
* [https://www.mediawiki.org/wiki/Special:MyLanguage/Localisation#Translation_resources Localise MediaWiki for your language] | |||
* | * [https://www.mediawiki.org/wiki/Special:MyLanguage/Manual:Combating_spam Learn how to combat spam on your wiki] | ||
* | * Consult the [[mediawikiwiki:Special:MyLanguage/Help:Contents|User's Guide]] for information on using the wiki software. | ||
* | |||
Latest revision as of 19:48, 9 January 2026
Weigang Qiu, Ph.D., Professor, Department of Biological Sciences Hunter College of City University of New York 413 East 69th Street, New York, NY 10021 Office: 1-212-896-0445 Email: wqiu-at-(hunter.cuny.edu) |
Fieldwork Gallery
Lab publications
Lyme Genomics, Evolution, & Ecology
Akther et al. 2024. "Natural selection and recombination at host-interacting lipoprotein loci drive genome diversification of Lyme disease and related bacteria" mBio 0:e01749-24. Press coverage: CUNY Graduate Center News Story; Hunter press
Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". The ISME Journal. 16, 447-464. (*co-first authors) Blog Post
Evolution & Learning Algorithms
Informatics Tool Development
last update: March 20, 2023
Lab members and trainees
| Year/Period | Doctoral members & trainees | Other members & trainees |
|---|---|---|
| Current Academic Year
(Fall 2022, Spring 2023 & Summer 2023) |
|
|
| Alumni (Since Fall 2002) |
|
(published coauthors)
|
Web Apps
Apps with Collaborators (or from published papers)
Borrelia diagnostic antigens (App developed by Liann Aris-Henry; Data from Arumugam et al (2019))
Borrelia growth transcriptome (App developed by Laziz Asamov; Data from Arnold et al (2016))
Data & GSEA associated with Polotskaia_etal_2014 (with Bargonetti Lab @Hunter)
Qiu Lab Apps
Curricular Development & Bioinformatics/QuBi Advising
- QuBi advisors: Weigang Qiu, Ntino Krampis, Rabindra Mandal (Biology); Saad Mneimeih, Lei Xie (CS); Akira Kawamura (Chem); Dana Sylvan (Math & Stats)
- Permission for non-Biology majors to take BIOL203 & BIOL425, every Spring
- Collect names, major, and IDs to send to course coordinator to grant permission. Waive BIOL10200 pre-reqs for taking BIOL203.
- Curricular resources:
- Biology courses and pre-reqs
- Hunter Biology Major 1 (including the Bioinformatics Option)
- Hunter Chemistry Major 2 (including the Bioinformatics Option)
- Hunter Computer Science (including Bioinformatics Concentration)
- Hunter Mathematics (including the Quantitative Biology Concentration)
- Hunter Statistics (including the Quantitative Biology Concentration)
- QuBi advising:
- Declaration of Bioinformatics concentration: In-person advising to work out the semester-by-semester courses
- Approve on department spreadsheet (or send email to "Samantha Sheppard-Lahiji" and "HTR Bio" <biology@hunter.cuny.edu>)
- Students should take Bioinformatics-specific electives (8 cred; see Hunter Catalog below), not general electives
- Examples: Anthrop302 (3 cr); Chem333 (3 cr); BIOL47119 & BIOL47120 (3cr); BIOL48002 (2 cr)
- Students need to take BIOL48002 (2 cr), which counts towards as research credit, to graduate as honors
- General advising:
- ~40 students every semester. Send out emails to students. Go through student courses by Email or by appointment
- Recommend new math courses: MATH15200 & STAT21350
- Hosting QuBi students in lab
- This is to enhance the informatics and coding skills of our students
- Students should register and get BIOL48002 credits, which counts towards their elective credits & eligibility for honors
- 3-5 students per semester
- Outside research opportunities
- MIT Quantitative Workshop (first week of January, in Boston). Coordination with CS (Saad & Susan Epstein) in Fall
- Simons Foundation/Flatiron Institute Center for Computational Biology (CCB) Internship program. Open House in Spring
Course/Lecture syllabi
- NYRaMP Workshop (August 2025, by Brandon Ely)
- Computational Genomics (KIZ, Fall 2024)
- NYRaMP Workshop (August 2024)
- BIOL47120 BioMedical Genomics (Spring 2024). Tutorials: R Markdown Cluster analysis single-cell RNA-seq
- BIOL425 Computational Molecular Biology (Spring, 2023). Lecture material on github
- BIOL714 Cell Biology: R Demo (Spring 2024) R Demo (Spring 2023)
- QuBi module: BIOL20300 Molecular Genetics, Lab 12 (2023)
- QuBi module: BIOL20300 Molecular Genetics, Lab 13 (2022)
- QuBi module: BIOL30300 Cell Biology, Bioinformatics Lab (transcriptome analysis)
- Big Data (Summer, 2020)
- BIOL47120 Biomedical Genomics II (Spring, 2020)
- BIOL425 Computational Molecular Biology (Spring, 2020)
- BIOL37500, Molecular Evolution (Fall, 2019)
- Southwest University R course (Summer, 2019)
- Analysis of Biological Data (Spring, 2017)
- Bioinformatics Workshop (Summer, 2014)
SARS-CoV-2 genome evolution
Akther, Bezrucenkovas, Sulkow, Panlasigui, Qiu, Di (April, 2020). "CoV Genome Tracker: tracing genomic footprints of Covid-19 pandemic". BioRxiv; Web app: SARS-CoV-2 Genome Tracker; Github: https://github.com/weigangq/cov-browser
Akther, Bezrucenlovas, Li, Sulkow, Di, Pante, Martin, Luft, Qiu (Sep, 2021). "Following the Trail of One Million Genomes: Footprints of SARS-CoV-2 Adaptation to Humans". BioRxiv link: . Github: https://github.com/weigangq/cov-db
Lab Resources & Protocols
- How to think like a computer scientist: An interactive Python programming book
- Nanopore sequencing protocols
- Monte Carlo Club
- NY-RaMP Mentoring
- Borreliella genome sequencing consortium: Weekly meetings (Tu @11): Since Jan 2023
- Borreliella diagnostic antigens (Fall 2023-Fall 2027):
- Zoom call (Jan 23, 2024)
- Next meeting: March 23, 2024
- Qiu lab network first-time user guide
- Qiu lab Github repositories: https://github.com/weigangq/?tab=repositories
- Mini-Protocols (frequently used computer codes and pipelines)
- Python tutorial: https://wiki.genometracker.org/~weigang/Intro_to_Python.html
- ick handling protocols
- Hunter HPC Usage
- R for Data Science (2e), (2024) by Wickham, Grolemund & Çetinkaya-Rundel (Bookdown version)
- Borrelia Genome Consortium: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA431102/
- Canadian Bbsl genome assemblies: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1130942
- Nanopore sequencing resources:
Wiki Help
- Configuration settings list
- MediaWiki FAQ
- MediaWiki release mailing list
- Localise MediaWiki for your language
- Learn how to combat spam on your wiki
- Consult the User's Guide for information on using the wiki software.