QuBi/modules/biol303: Difference between revisions
imported>Weigang (Created page with "<span style="color: Seagreen;font-weight:bold;font-size:large;">Bioinformatics Lab: Exploration of Gene Expression in ''Dictyostelium species''<span> ==Objectives== # Und...") |
imported>Weigang mNo edit summary |
||
| Line 1: | Line 1: | ||
<span style="color: Seagreen;font-weight:bold;font-size:large;">Bioinformatics Lab: Exploration of Gene Expression in ''Dictyostelium | <span style="color: Seagreen;font-weight:bold;font-size:large;">Bioinformatics Lab: Exploration of Gene Expression in ''Dictyostelium'' species<span> | ||
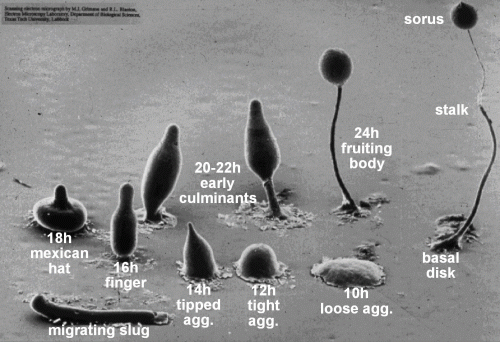
[[File:Dicty-cycle.gif|thumbnail|left|Development of Dictyostelium (M. Grimson, R. Blanton, Texas Tech University)]] | |||
==Objectives== | ==Objectives== | ||
Revision as of 01:01, 27 August 2014
Bioinformatics Lab: Exploration of Gene Expression in Dictyostelium species
Objectives
- Understand the microarray technology and its use in genomewide identification of gene functions.
- Be able to identify coexpressed and corepressed genes based on timecourse gene expression data.
Lab Report Grading Policy
- Introduction (3pt)
- Define transcriptome. Describe advantages of highthroughput technologies in comparison with traditional genebygene approaches of studying gene function. Your statements are not to be copied from the Lab Manual.
- Materials and Methods (4pts)
- Define “diauxic shift”. Summarize the design of the DNA chip. Describe experimental procedures of the DeRisi study that have produced these gene expression data.
- Results (12pts)
- Answers to twelve questions A L. Reproduce answers from your worksheets. For the questions that are TABLES, copy the table but you can omit some rows as indicated on some of the table captions.
- Discussion (9pts)
- Answer three discussion questions.
- Summary/Conclusion (1pt)
- A sentence or two will suffice.
- References (1pt)
- Credit is given for pertinent references obtained from sources other than the Lab Manual.
Introduction
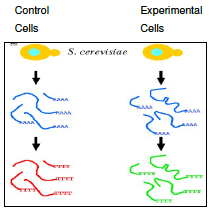
Gene expression is the transcription of a DNA template into RNA molecules. In a multicellular organism, the subset of genes that are expressed defines and gives rise to a specific tissue or cell type. In a single-celled organism (such as Figure 1, Saccharomyces cerevisiae, the baker’s yeast), genes are turned on and off depending on environmental conditions. In this laboratory exercise, we will use bioinformatics techniques to identity genes that are expressed in the yeast during glucose starvation.
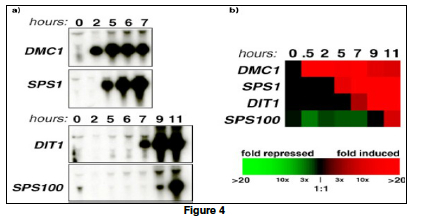
Traditionally, gene expressions are studied one gene at a time using blotting techniques. For example, in a Northern Blot experiment (Figure 4a), the whole messenger RNA (mRNA) content of a cell is extracted and loaded on a solid gel slab. Different mRNA molecules are then separated using electrophoresis and transferred to a nitrocellulose sheet. To identify if a gene is expressed, a radioactively (or fluorescently) labeled oligonucleotide probe that is specific to the gene sequence is applied to the sheet. If the gene is expressed, the probe will hybridize with a specific mRNA molecule and a black band will appear on an Xray film. Other blotting techniques for detecting gene expression include Southern Blot, in which mRNAs in a cell are reverse transcribed to their complementary DNA (cDNA) before being hybridized with gene-specific oligo-nucleotide probes. In a Western Blot experiment, the protein product (instead of the mRNA intermediate) of a gene is probed using antibodies (instead of the oligonucleotide probes).
After the genomic revolution since 1990s, it became possible to study the expression of all genes in a cell at once using high-throughput techniques. Detecting the expression profiles of a whole genome was made possible by the availability of the whole genome sequences of bacteria, yeasts, and humans. The DNA microarray is one such high throughput technique. In contrast to the Northern Blot technique in which the mRNA sample is fixed on a nylon sheet, nucleotide probes for all genes are fixed on a glass slide, creating a “gene chip”. The cellular mRNAs are reverse transcribed into cDNAs labeled with fluorescent dyes, which are then hybridized with the gene chips. After the unattached cDNAs are washed away, the fluorescent intensity remains at each probe location is measured as an indication of the amount of mRNA transcribed from each gene in a genome. The entire cellular RNA content transcribed from a genome is called a transcriptome. Each DNA microarray reading is therefore essentially a snap shot of the whole genome expression profile of a cell at a particular physiological stage. It is no longer necessary to know or decide beforehand candidate genes to be targets of exploration, as in the traditional blotting techniques.
Most recently, direct sequencing of the whole mRNA content of a cell using the so-called “next-generation” sequencing technologies provides an alternative and even more accurate way of obtaining the transcriptome of a cell. These high-throughput technologies, however, create new technical challenges of their own. The main challenge is the analysis of the huge amount of data resulting from each microarray or sequencing experiment. First, data from high-throughput experiments need computer-assisted data processing and analysis. Second, statistical analysis and testing become essential tools for the discovery and exploration of gene functions, e.g., finding co-expressed genes.
Procedures
1. Microarray Color Prediction
Question A: Consider a microarray with only two spots, one for Gene A and one for Gene B. A researcher cultures two yeast strains in Glucose solution, extracts mRNA from the cells, creates cDNA with either red dye (for the experimental strain) or green dye (for the control strain). The dye containing cDNA is then allowed to combine with a microarray containing spots with complementary DNA for Gene A and Gene B. In each situation below, predict what color the two spots will show. The first answer is filled in for you as an example.
Question B: Now consider a culture where one yeast population (the experimental group) is grown in a solution where the amount of glucose decreases over time to zero, while a second population (the control group) is grown in flask where the Glucose concentration remains nearly constant. The researcher then extracts mRNA from the yeast cells to creates red-dyed-DNA for the experimental strain and green dye for cDNA made from the control strain. The dye-containing cDNA is then allowed to combine with a microarray containing spots with complementary DNA for a cytochrome C gene and a tRNA synthesase Gene. The expression levels in the control group are assumed to remain constant for both genes but the expression levels for the experimental group change according to the graph in Figure 5. In each situation, predict what color the two spots will show.
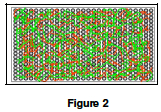
2. Examining an Array Image

Question C: For this question you will examine the top three rows from a microarray. Open a browser and access the image at http://cmgm.stanford.edu/pbrown/explore/M6.jpg. This is the magnified scan of an actual microarray from glucose deprived yeast. If you cannot access the image you may use an image provided by your lab instructor or Color Plate Figure 3.
3. Quantifying the Array Color Intensity
In microarray analysis we need to precisely quantify the level of gene expression. The machine measured brightness of red and yellow dye is usually put into a table and expressed as a fold number. Fold numbers are a measure of doubling and are the base-2-logarithm of the color signal intensity ratios.
The first line of the chart below is not from the microarray, it is simply a measurement of colony growth taken by shining light through the yeast colony as the cells multiplied. The second line is a measure of the ever-dropping nutrient concentration in the medium.
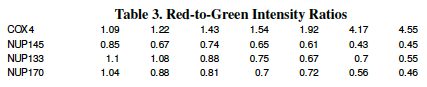
The third and fourth lines quantify the color of a single microarray spot (in this case, the gene NUP120) over time. For each time point, calculate the ratio of color intensity (red / green) and the base-two-logarithm of that ratio [ log2(red/green) ] . The first has been done for you as an example.
Question D: On your lab report only copy the portions of Table 2 which you filled in, i.e. the bottom two rows. Your answers may differ slightly from your labmates, depending on your perception of color and quality of the image you are viewing. Table 1 is only meant to be an estimation, not an exact number.
Question E: What can you tell about the yeast cell density by looking at the O.D. line of the chart? How was this measured?
Question F: What molecule must bind to the microarray in order to create a strong green signal? What makes the green glow? Which type of cells, the control or the experimental sample, is the green signal associated with and which type of cells is the red signal associated with?
- Quick review of logarithms:
- Base two logarithms are a measure of how much a thing has doubled. log2(1) = 0, log2(8) = 3, log2 of (.25) = 2 [note the negative sign]. If your calculator doesn't have a log2 function, you may use log10 and multiply your result by 3.3219.
4. Explore the Expression Profile using EXCEL
The data in Table 3 (Red-to-Green Intensity Ratios) were measured in an experiment where two strains of yeast were grown under conditions of decreasing glucose concentration. A whole-genome microarray measured the expression levels of 6,400 genes for the two strains as the glucose level in the medium decreased. Data for 4 of those genes are shown below; the ratio of red fluorescence intensity to green fluorescence intensity for 4 genes (COX14, NUP145, NUP133 and NUP170) are shown.
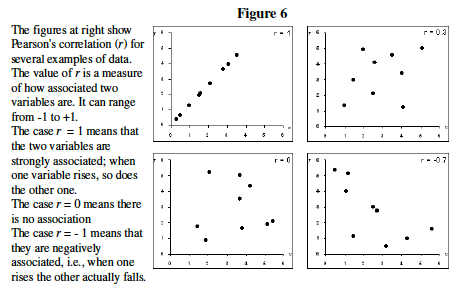
- By just looking at the Figure 6, make your best guess, which two genes below seem to have the most similar expression levels ? ___________ and _______.
- Guessing is not the best way to describe gene clusters. Statistics has a variety of ways to measure correlation. We will use Pearson's Correlation Coefficient, r.
- Excel easily measures correlation and Pearson's r. Like most calculations in Excel, you simply click on an empty cell, type "=", write a formula, and indicate what range of cells you wish to perform the calculation on.
The above example was created by clicking on Cell J3 and entering the formula: =CORREL(B1:H1,B2:H2 ) which caused the spreadsheet to compare the values in Row 1 and Row 2 and print a correlation value (Pearson's r) in Cell J3 .
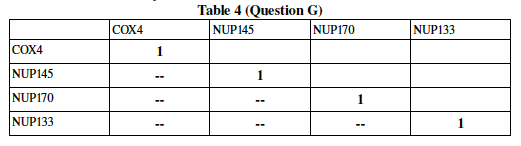
You will do this for six different pairs (one correlation has to be measured for every possible twosome, i.e. first and second row, first and third row, etc) Write your six pairwise correlation results into the following table. The correlation of any gene with itself is, of course, perfect, and hence "1". A double dash has been placed in half of the spaces to save you the trouble of writing a result twice. This may take some time, especially if you are unfamiliar with Excel, spreadsheets, or Statistics. Work together and ask someone who can help.
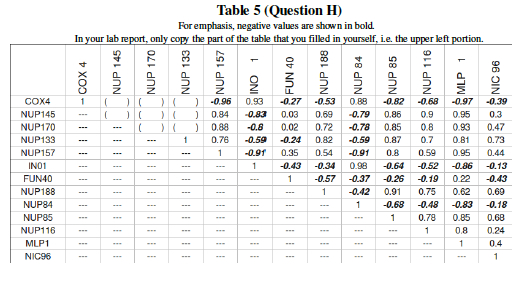
Now, since your data is from a much larger experiment (DeRisi et al measured all 6000plus genes in the Yeast genome), transfer your numbers into a bigger table. The table below describes the pairwise correlation for 13 yeast genes undergoing glucose deprivation. Add your correlation data calculated in the previous question to the 6 blank spaces in Table 5.
Question I: Name two pairs of genes which act in tandem, i.e. rise together and fall together as the yeast cell experiences glucose deprivation.
Question J: Name two pairs of genes where, if one gene is over-expressed, the other gene will be suppressed.
5. Database Search
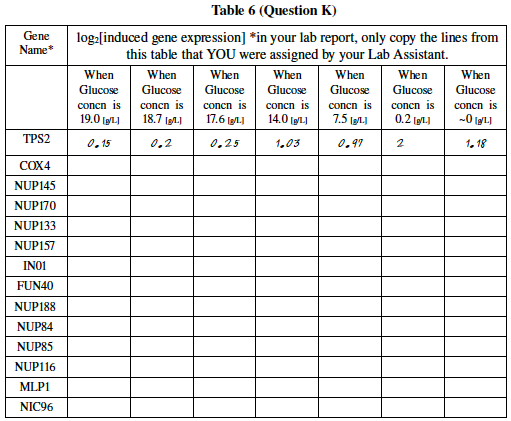
Your lab instructor. will assign a few genes below to you or to your group. For whichever genes your lab instructor has assigned to you, do a fold calculation using the same math you employed in Step 2, where you calculated the ratio of gene expression (level in the experimental group divided by level in the control group). Recall that you then found the base two log (log2) of this ratio. Starving yeast cell may be expected to alter the enzymes in its metabolic pathway.
Get your data by searching for your gene name at http://cmgm.stanford.edu/pbrown/explore/diauxsearch.html. Use the buttons that allow you to put in a Description keyword. Note that, although the website data is labeled as a fold it is actually a simple ratio of the two colors, so you will need to take that fold and take the base 2 log of it. A line of correct answers for TPS2 has been written in for you as an example.
- *For example, the DeRisi experiment found that the fold increase of TPS2 was 1.11, 1.15, 1.19, 2.04, 1.96, 4, 2.27. From this one can calculate the log2 values filled into the first row of Table 6.
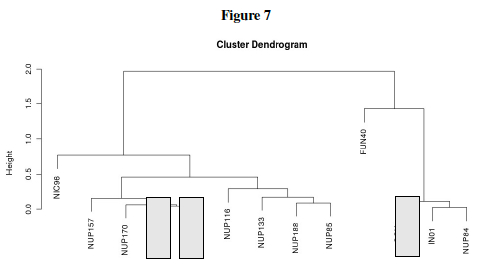
6. Distance Matrix and Clustering Dendrogram
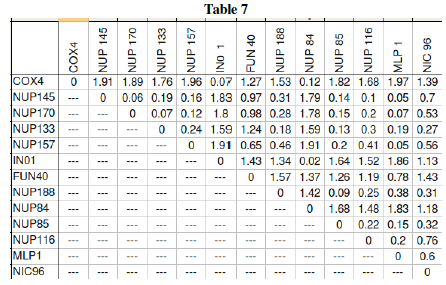
Question L: Much of the previous questions in this lab were just preliminary work leading up to one big question: Which genes form interactive networks? Let us now attempt to answer that! Table 5 showed the behavior of yeast genes under glucose deprivation. The correlation data (Pearson's correlation, r) can are interpreted in Table 7 as distances. The Table 7 data shows large values for disparate genes and approaches 0 for the ideal case where two genes are expressed in exactly the same levels in varying conditions. This then can be graphically shown as a dendrogram (tree) where similar expression gives close tree-distance. Three genes: NUP 145, COX4, and MLP1 still need to be placed on the dendrogram. Using the pairwise distances from Table 7, place these three genes in the proper location within the tree.
Discussion Questions
Question M: What differences and similarities does a DNA chip have with a Northern blot?
Question N: It is stated in the DeRisi paper that “[k]nowing when and where a gene is expressed often provides a strong clue as to its biological role”. Explain how a time-course experiment using microarray could be used for discovering genes in a metabolic network (e.g., glucose utilization), or in a subcellular structure (e.g., the nuclear pore complex).
Question O: In the next statement, the DeRisi paper writes that “Conversely, the pattern of genes expressed in a cell can provide detailed information about its state”. Describe how the whole-genome expression profiles could be used for, e.g., early diagnosis of cancer and drug discovery.
References & Resources
- DeRisi, Joseph.L. , Iyer, Vishwanath R., Brown, Patrick O. ; Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale, Science 24 October 1997:Vol. 278. no. 5338, pp. 680 686; DOI: 10.1126/science.278.5338.680. The article is online at: http://www.sciencemag.org/cgi/content/full/278/5338/680, accessed on Dec. 11, 2009.
- Most of the data in this lab activity was obtained from the extensive DeRisi lab website: http://cmgm.stanford.edu/pbrown/explore/index.html, accessed on Dec 11, 2009.
- Figures 1, 2, and 4 came from Campbell, A. M. and L. J. Heyer. (2003). Discovering Genomics, Proteomics, & Bioinformatics. Pearson Education, Inc. San Francisco, CA. Unit Two: Gene Expression
- Figure 3 came from the Pat Brown lab at Stanford Medical School : http://cmgm.stanford.edu/pbrown/explore/M6.jpg
- Zvelebil, M. & J. O. Baum. (2008). Understanding Bioinformatics. Garland Science. Chapter 16: Clustering Methods and Statistics.
--Dphilli (talk) 12:56, 6 August 2013 (EDT)
- Read this experimental report and extract the following information:
- Name of the two species used in experiments
- How many genes were measured for their expression (i.e., mRNA) levels?
- Describe a biological question that can be answered by this experiment (e.g., which genes are expressed at a particular developmental stage)
- Go to dictyExpress and explore the time course of a set of genes
- Choose the 2nd Box: "Run dictyExpress (RNA-seq)"
- In the "Gene Selection" Box, type the following gene names one at a time (DON'T copy and paste; when the gene is found, highlight it and press enter): acrA, catB, dcsA, acgA, abcG18
- Click "Update" and answer the question based on the plot in the "Expression Profile" panel: Are these genes up- or down-regulated during development?
- Do the same for the 2nd set of genes: mserS, rpl38, rpsA, rpl35a, gfm1
- Do the same for the 3rd set of genes: gefB, gefX, gxcB, mgp3, gefN
- Combine all 3 sets of genes and produce a heatmap
- In the "Hierarchical Clustering" Panel, choose the "Pearson Correlation" for "Distance Function"
- Choose "Average Linkage" for "Linkage" and your choice of color gradient
- What is represented by each row?
- What is represented by each column?
- Do these 3 sets of genes form clusters by themselves?
- HHMI slides: A technical description of how to group genes and samples by their overall similarity in gene expression levels