Main Page: Difference between revisions
| Line 89: | Line 89: | ||
==Web Apps== | ==Web Apps== | ||
=== | ===Apps with Collaborators=== | ||
<gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | <gallery mode="packed" heights="200px" perrow="4" style="text-align:left"> | ||
File:Screenshot_2023-03-24_122016.png| link=http://borreliabase.org/~wgqiu/clickme-khalikuz/temp-Points.html | Phosphoproteome MDM2KD data set (with Bargonetti Lab @Hunter) | File:Screenshot_2023-03-24_122016.png| link=http://borreliabase.org/~wgqiu/clickme-khalikuz/temp-Points.html | Phosphoproteome MDM2KD data set (with Bargonetti Lab @Hunter) | ||
Revision as of 19:43, 25 March 2023
Weigang Qiu, Ph.D., Professor Department of Biological Sciences Hunter College of City University of New York Belfer Research Building, Room 402 413 East 69th Street, New York, NY 10021 Office: 1-212-896-0445 Email: wqiu-at-(hunter.cuny.edu) Directions by Google Map |
Lab publications
Lyme Genomics
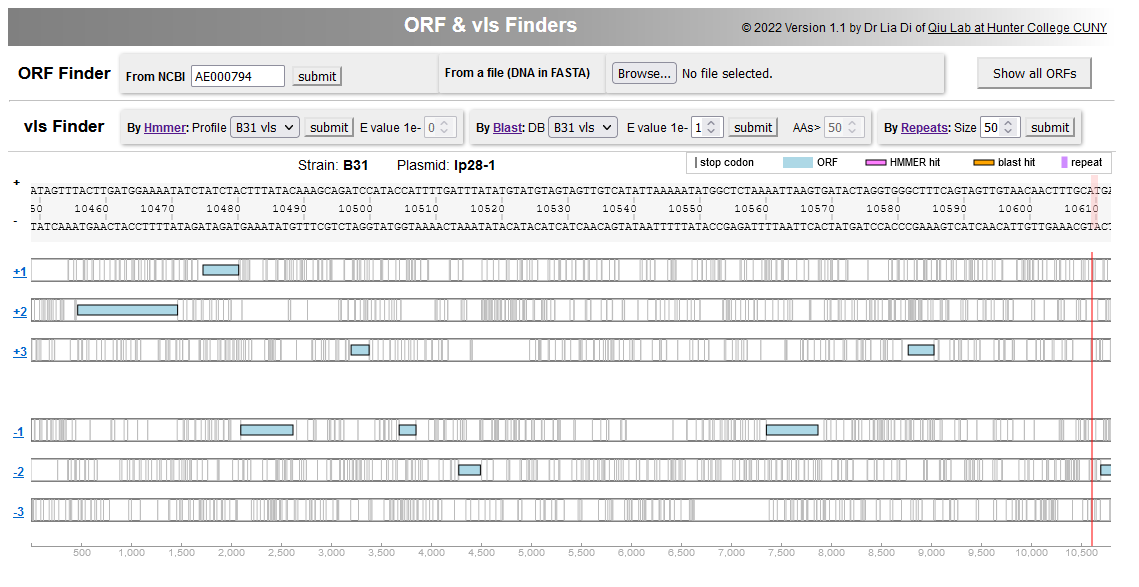
Li, Di, Zeglis, Qiu (2022). “Evolution of the vls antigenic variability locus of the Lyme Disease pathogen and development of recombinant monoclonal antibodies targeting conserved VlsE epitopes”. Microbial Spectrum 10 (5):1-15. DOI: https://doi.org/10.1128/spectrum.01743-22
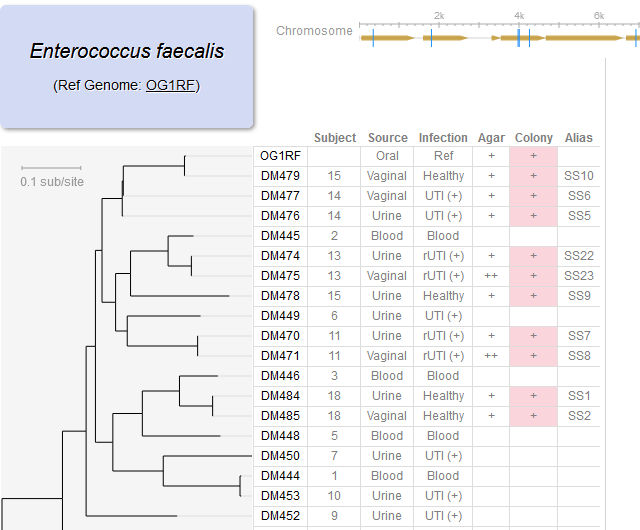
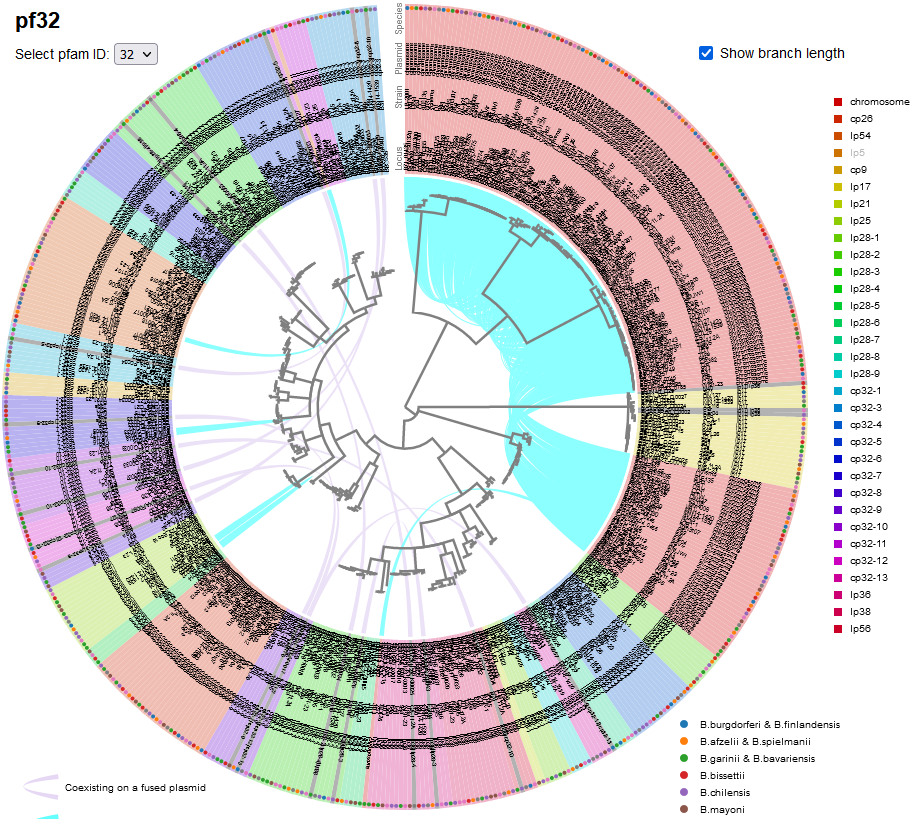
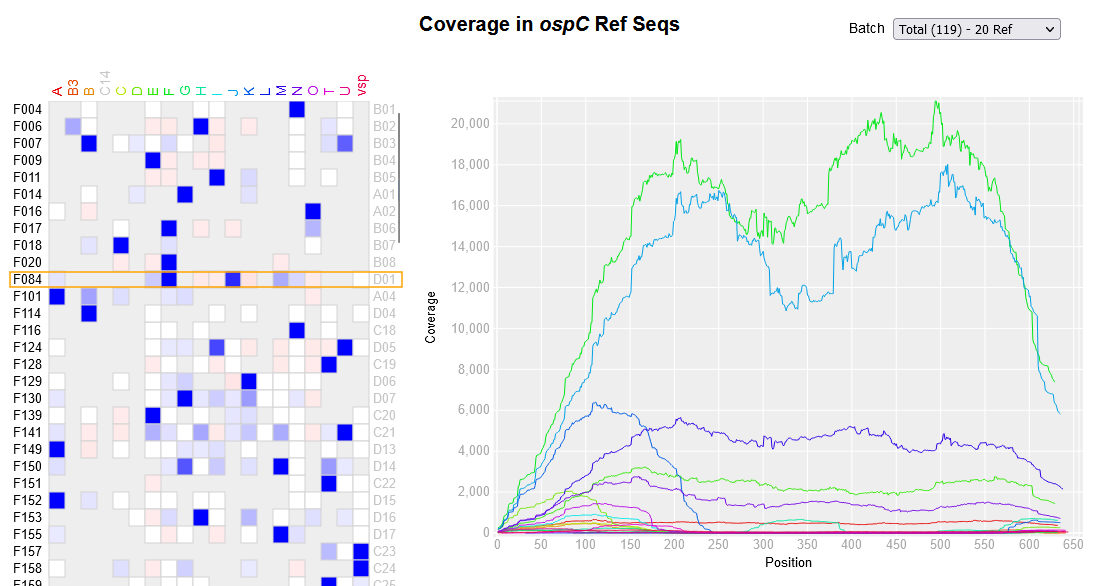
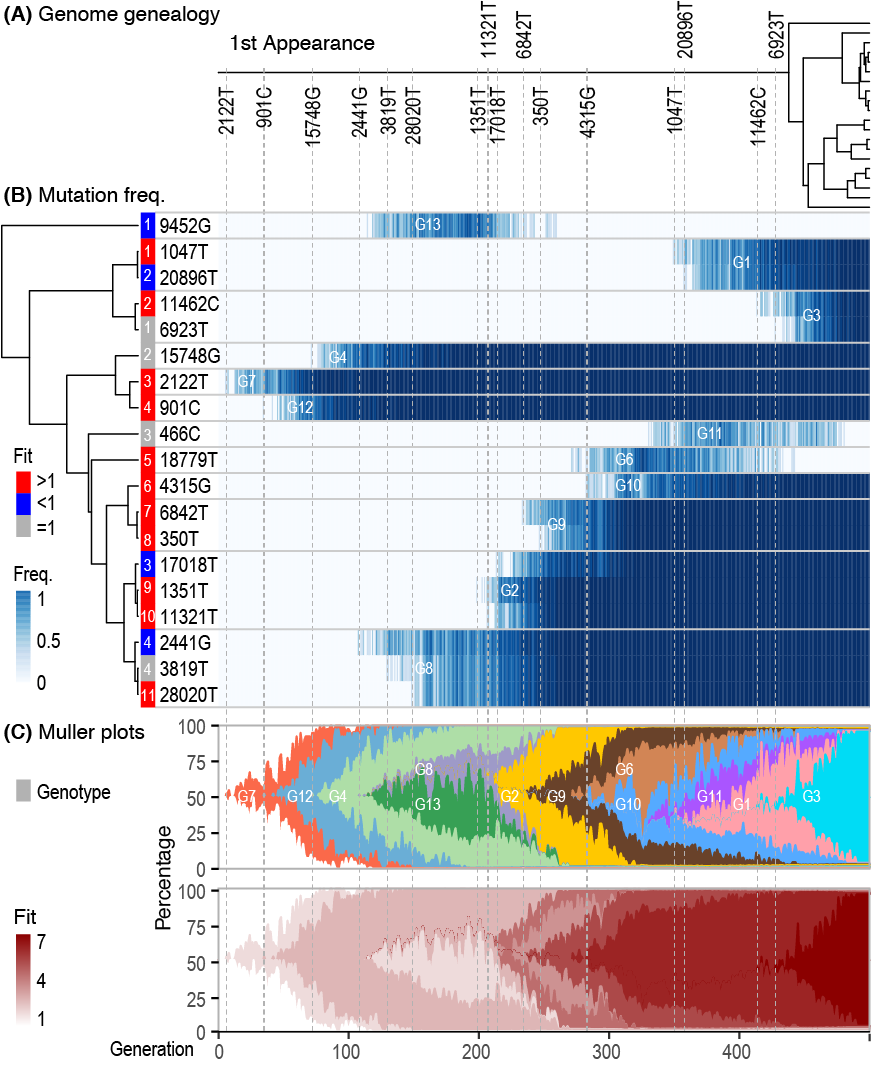
Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". The ISME Journal. 16, 447-464. [PMID 34413477] (*co-first authors)
Schwartz, Margos, Casjens, Qiu, Eggers (2020). "Multipartite Genome of Lyme Disease Borrelia: Structure, Variation and Prophages". Current Issues in Molecular Biology. 42:409-454. DOI: https://doi.org/10.21775/9781913652616
Barbour & Qiu (2019). Borreliella. In Bergey's Manual of Systematics of Archaea and Bacteria (eds W. B. Whitman, F. Rainey, P. Kämpfer, M. Trujillo, J. Chun, P. DeVos, B. Hedlund and S. Dedysh). John Wiley & Sons, Inc., in association with Bergey's Manual Trust. DOI: https://doi.org/10.1002/9781118960608.gbm01525
Evolution & Learning Algorithms
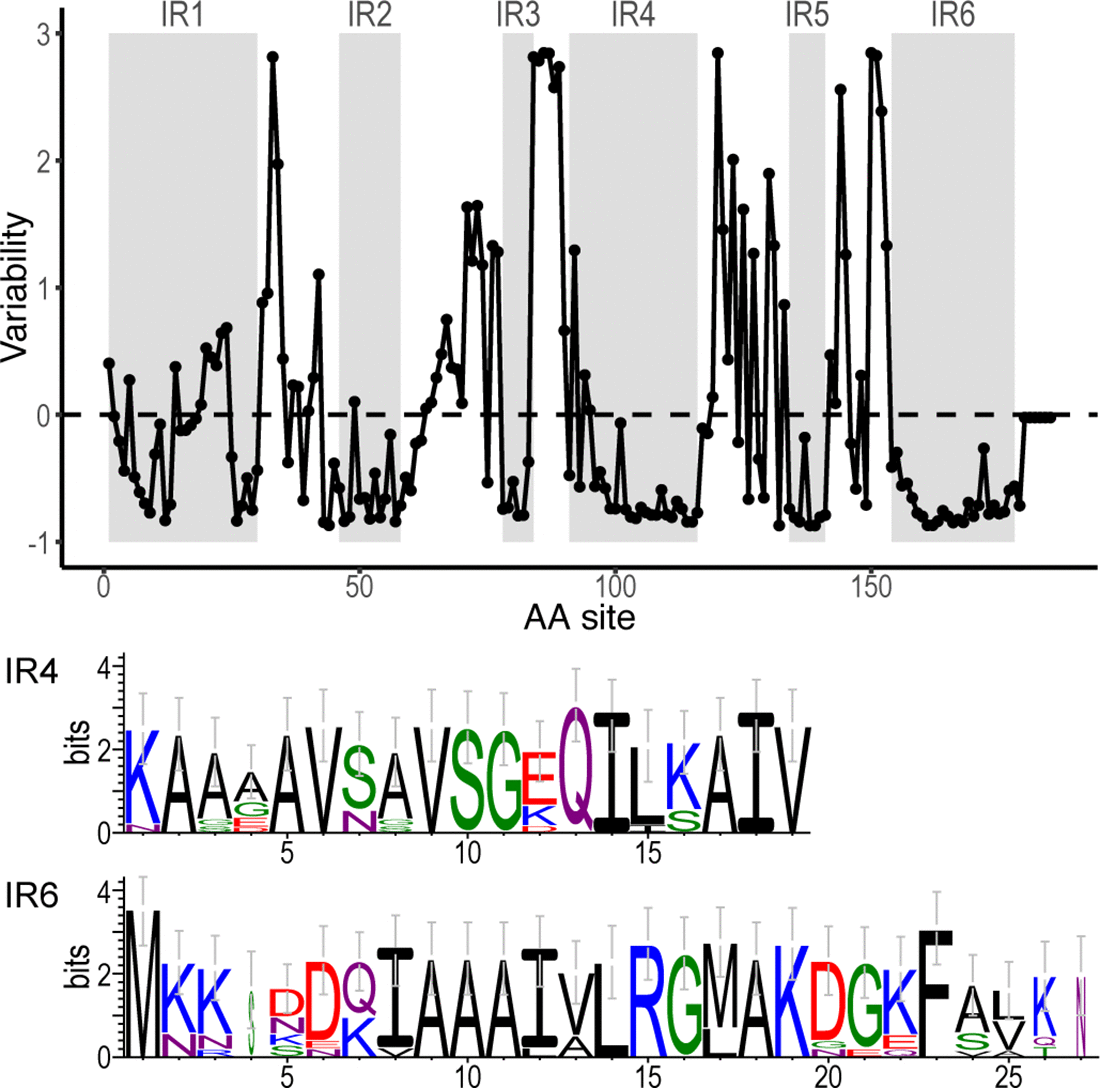
Ely, Koh, Ho, Hassan, Pham, Qiu (2023). Novelty Search Promotes Antigenic Diversity in Microbial Pathogens. Pathogens 12:388. DOI: https://doi.org/10.3390/pathogens12030388
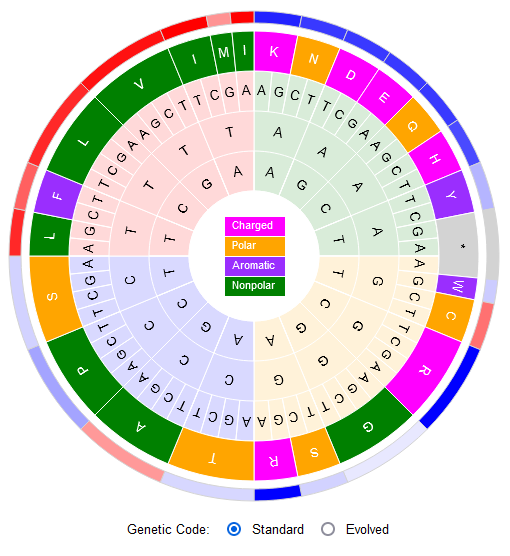
Attie, Sulkow, Di, Qiu (2019). Genetic codes optimized as a traveling salesman problem. PLoS ONE 14(10): e0224552. DOI: https://doi.org/10.1371/journal.pone.0224552
Yan, Deforet, Boyle, Rahman, Liang, Okegbe, Dietrich, Qiu, Xavier (2017). Bow-tie signaling in c-di-GMP: Machine learning in a simple biochemical network. PLoS Comput Biol 13(8): e1005677. DOI: https://doi.org/10.1371/journal.pcbi.1005677
Informatics Tool Development
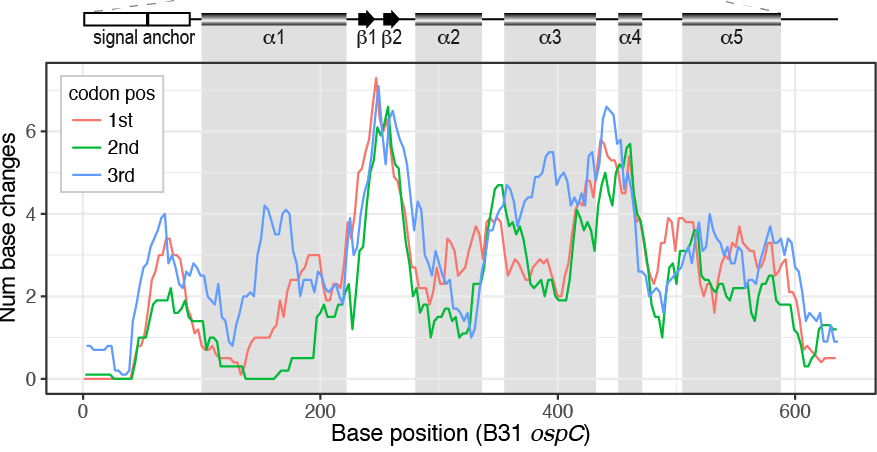
Hernandez, Bernstein, Pagan, Vargas, McCaig, Ramrattan, Akther, Larracuente, Di, Vieira, Qiu. (2018). BbWrapper: BioPerl-based sequence and tree utilities for rapid prototyping of bioinformatics pipelines. BMC Bioinformatics 19(1):76. [PMID 29499649]
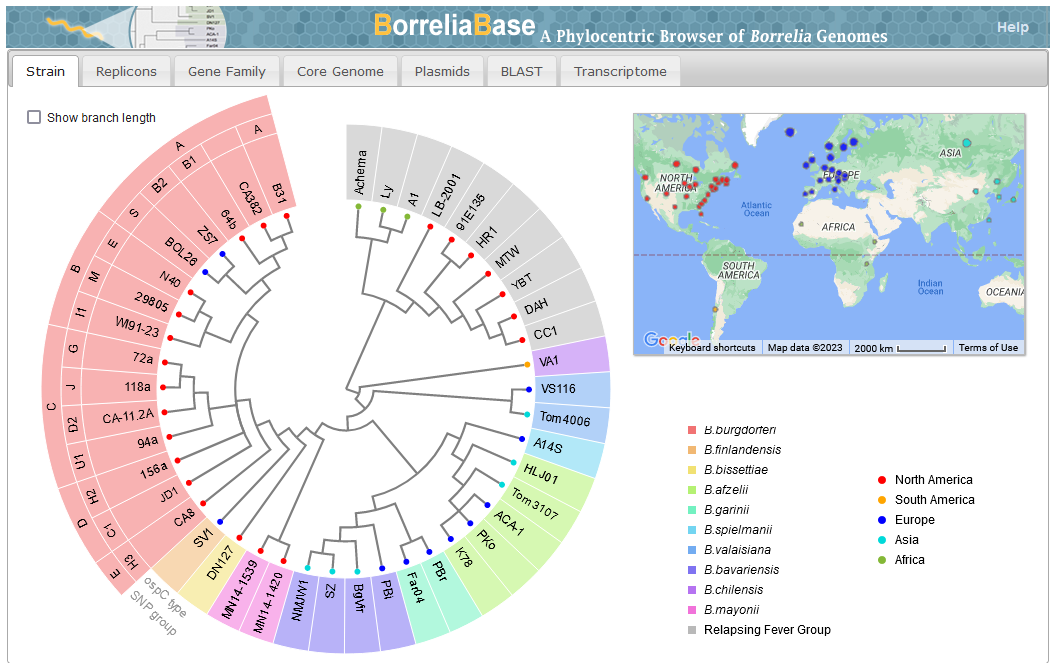
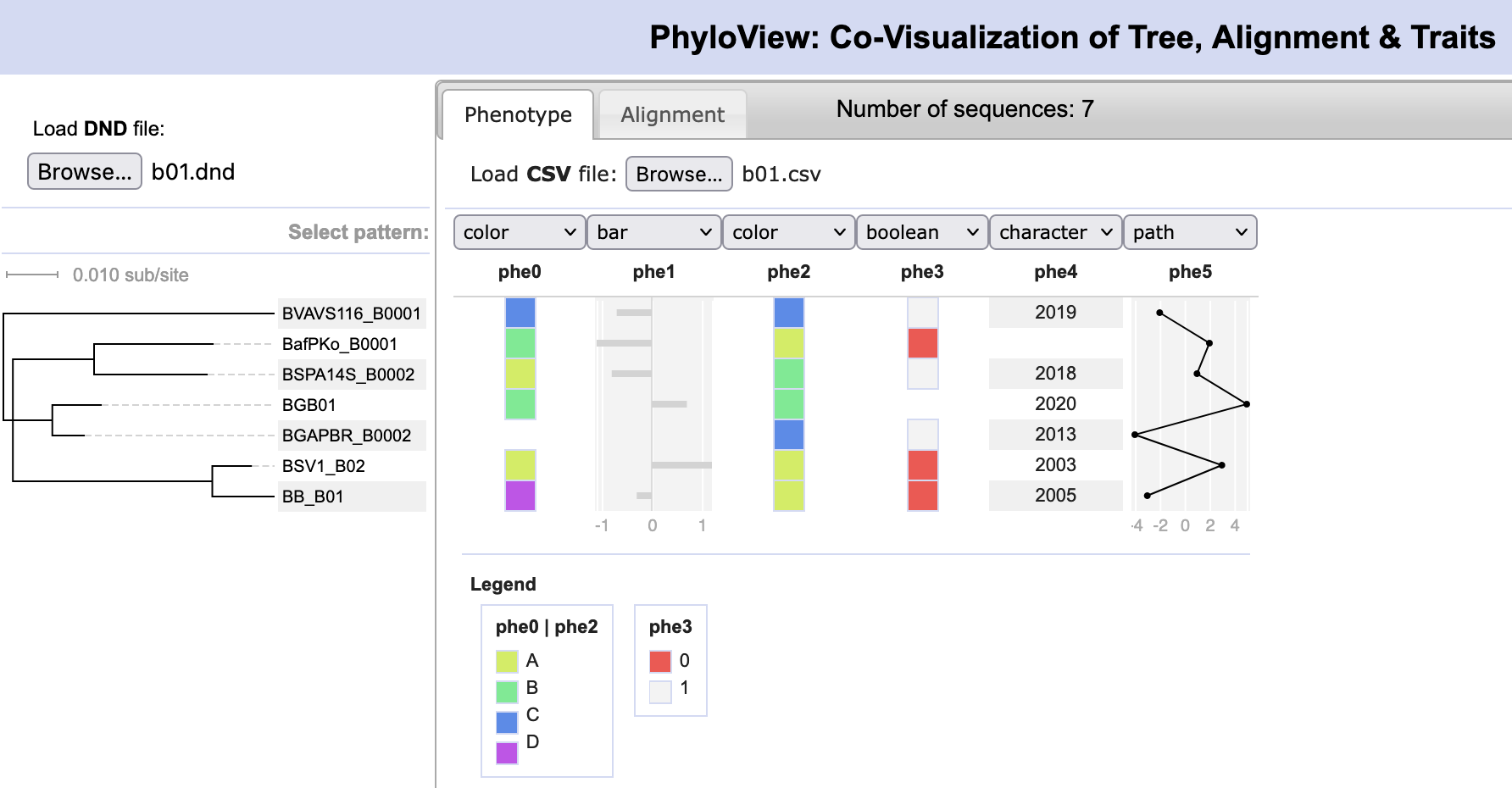
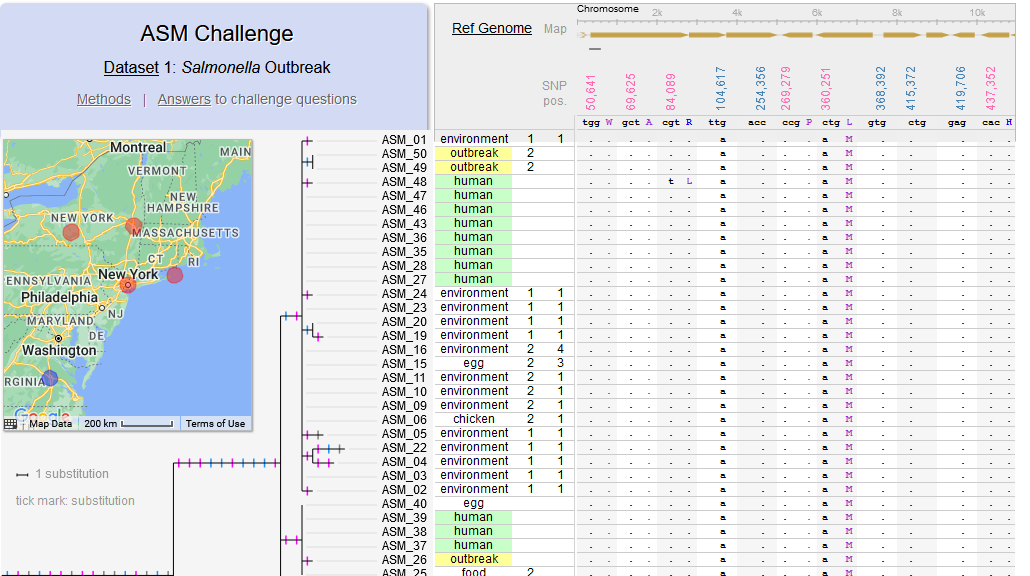
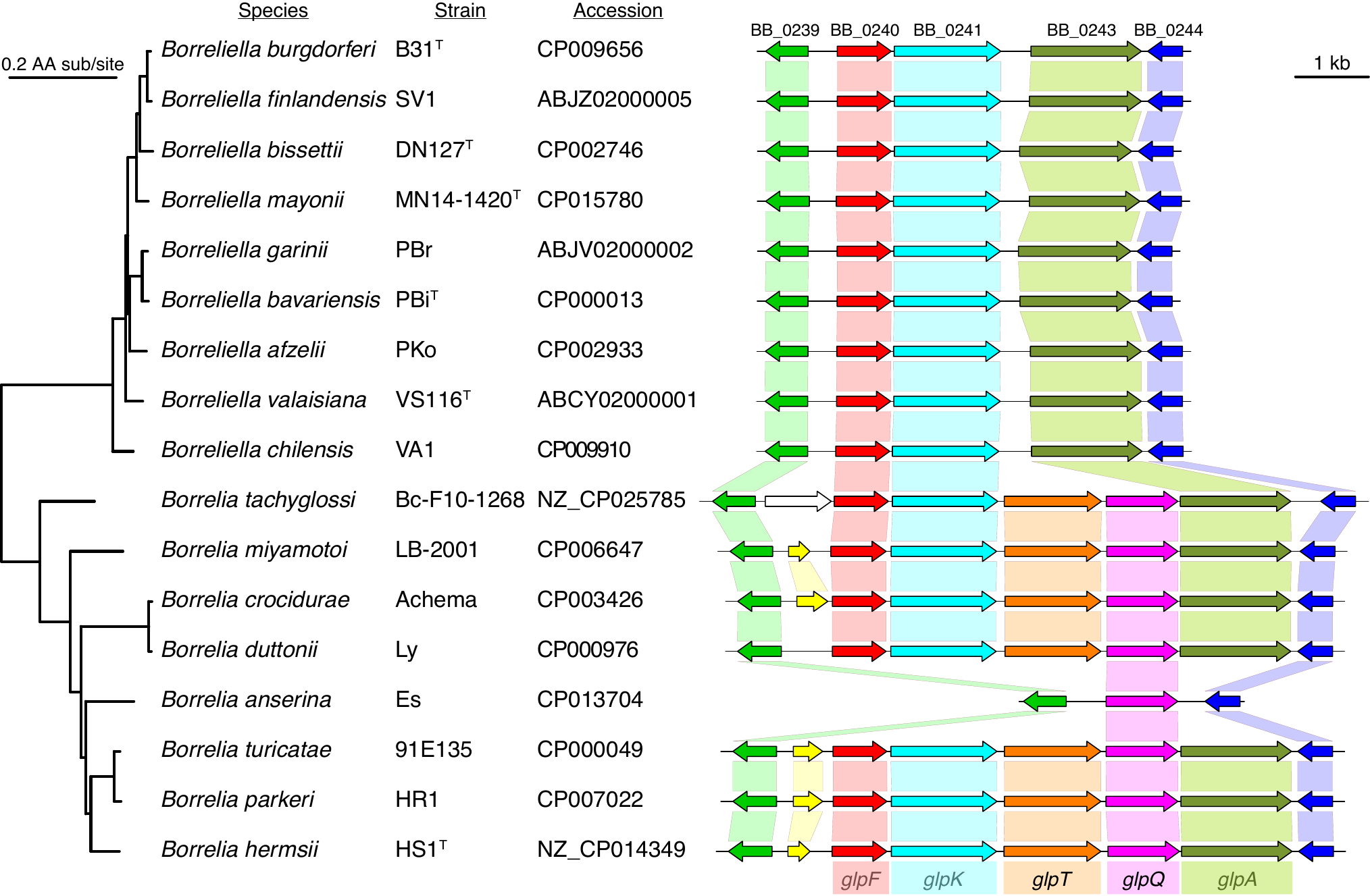
Di, Pagan, Packer, Martin, Akther, Ramrattan, Mongodin, Fraser, Schutzer, Luft, Casjens and Qiu. (2014). BorreliaBase: a phylogeny-centered browser of Borrelia genomes. BMC Bioinformatics 15:233. [PMID 24994456]
last update: March 20, 2023
Lab members and trainees
| Year/Period | Doctoral members & trainees | Other members & trainees |
|---|---|---|
| Current Academic Year
(Fall 2022, Spring 2023 & Summer 2023) |
|
|
| Alumni (Since Fall 2002) |
|
(published coauthors)
|
Web Apps
Apps with Collaborators
Qiu Lab Apps
Teaching & Curricular Development
- The Quantitative Biology (QuBi) Program at Hunter
- Hunter Biology Major I (including the Bioinformatics Conentration)
- Hunter Chemistry Major 2 (including the Bioinformatics Option)
- Hunter Computer Science (including Bioinformatics Concentration)
- Hunter Mathematics (including the Quantitative Biology Concentration)
- Hunter Statistics (including the Quantitative Biology Concentration)
- QuBi module: BIOL20300 Molecular Genetics, Lab 4
- QuBi module: BIOL20300 Molecular Genetics, Lab 7
- QuBi module: BIOL30300 Cell Biology, Bioinformatics Lab (transcriptome analysis)
- Big Data (2020)
- Southwest University R course (2019)
- BIOL425 Computational Molecular Biology (2020)
- Analysis of Biological Data (2017)
- BIOL37500, Molecular Evolution (2019)
- Bioinformatics Workshop (2014)
- BIOL47120 Biomedical Genomics II (2020)
SARS-CoV-2 genome evolution
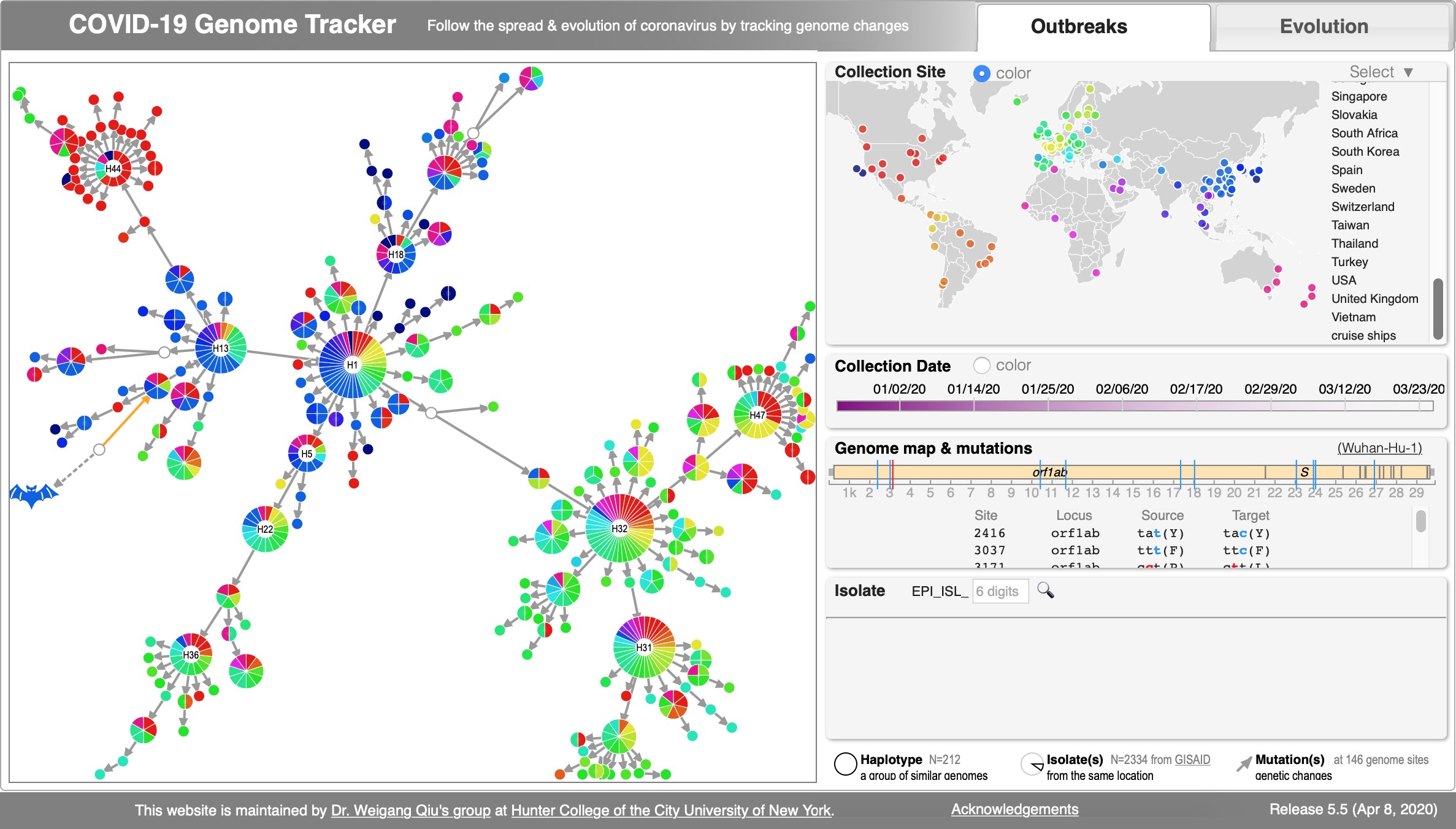
Akther, Bezrucenkovas, Sulkow, Panlasigui, Qiu, Di (April, 2020). "CoV Genome Tracker: tracing genomic footprints of Covid-19 pandemic". BioRxiv; Web app: SARS-CoV-2 Genome Tracker; Github: https://github.com/weigangq/cov-browser
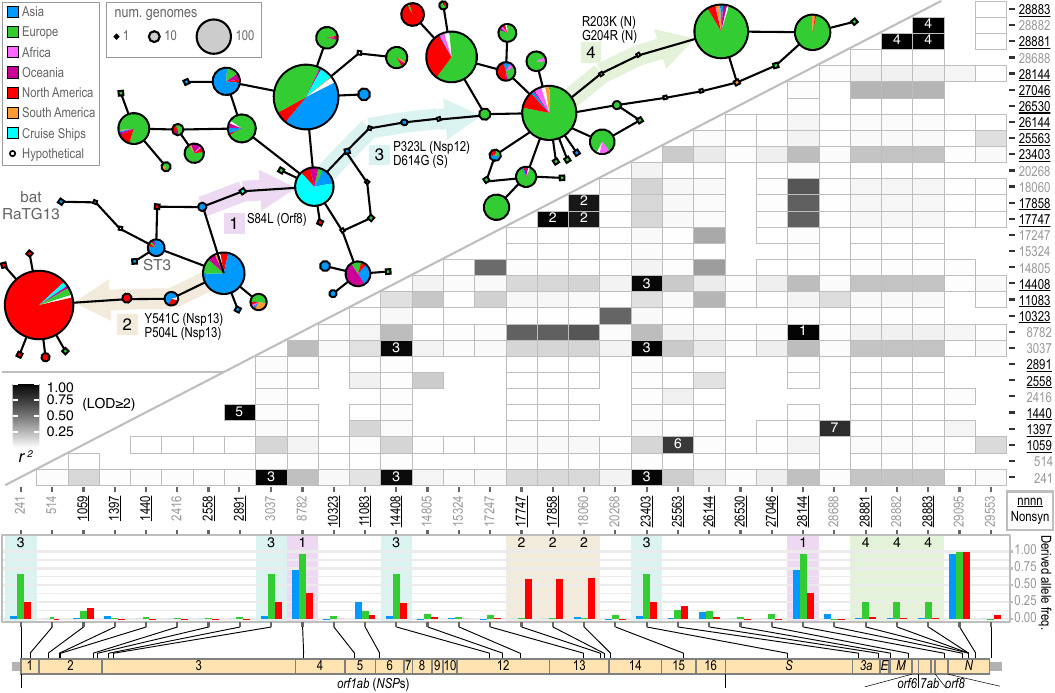
Akther, Bezrucenlovas, Li, Sulkow, Di, Pante, Martin, Luft, Qiu (Sep, 2021). "Following the Trail of One Million Genomes: Footprints of SARS-CoV-2 Adaptation to Humans". BioRxiv link: . Github: https://github.com/weigangq/cov-db
Lab Resources & Protocols
- Github repositories: https://github.com/weigangq/?tab=repositories
- Mini-Protocols (frequently used computer codes and pipelines)
- Monte Carlo Club (summer projects)
- Tick handling protocols
- Hunter HPC Usage
Wiki Help
- Configuration settings list
- MediaWiki FAQ
- MediaWiki release mailing list
- Localise MediaWiki for your language
- Learn how to combat spam on your wiki
- Consult the User's Guide for information on using the wiki software.


![Di*, Akther*, Bezrucenkovas, Ivanova, Sulkow, Wu, Mneimneh, Gomes-Solecki, Qiu (2021). "Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity". The ISME Journal. 16, 447-464. [PMID 34413477] (*co-first authors)](/images/9/93/Fig7-small.png)
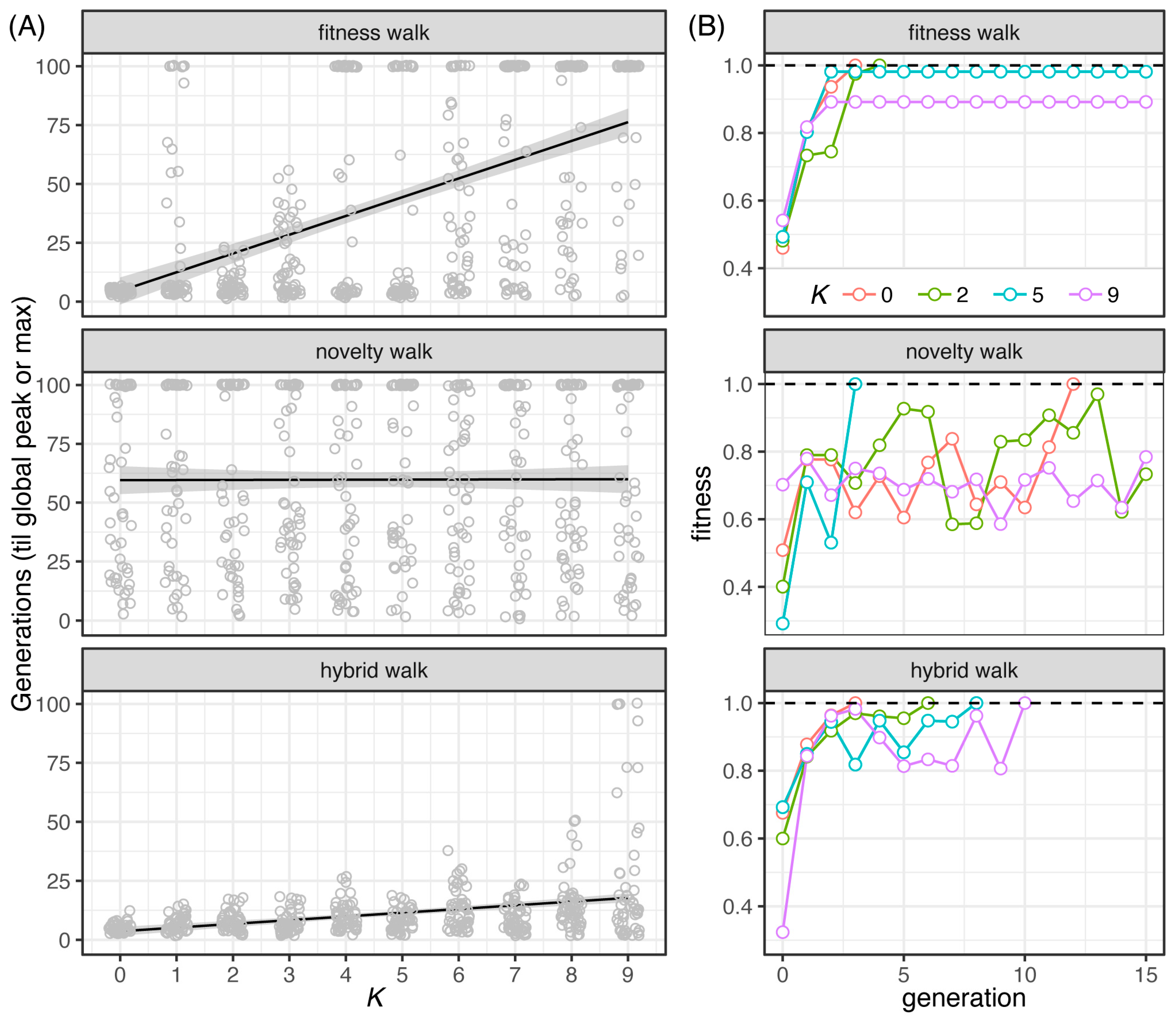
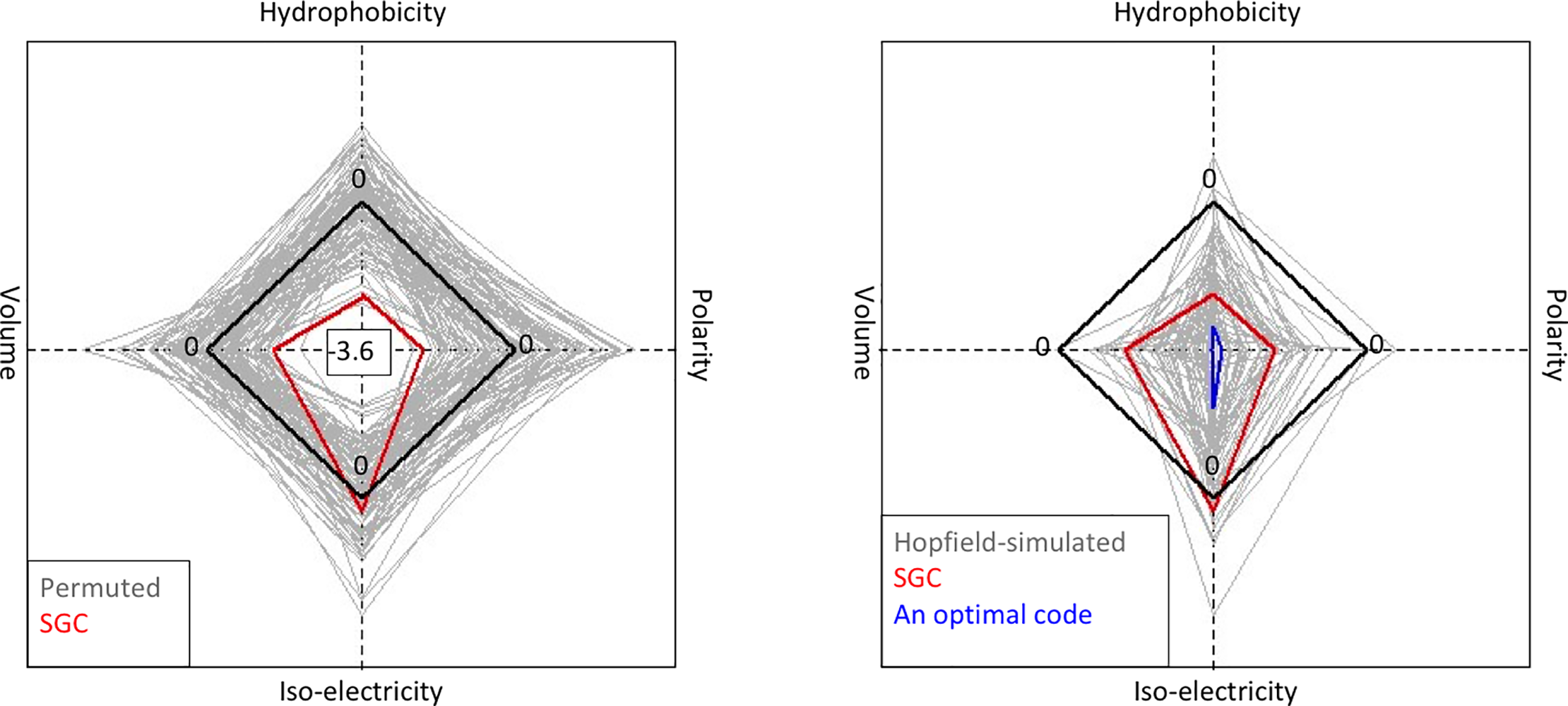
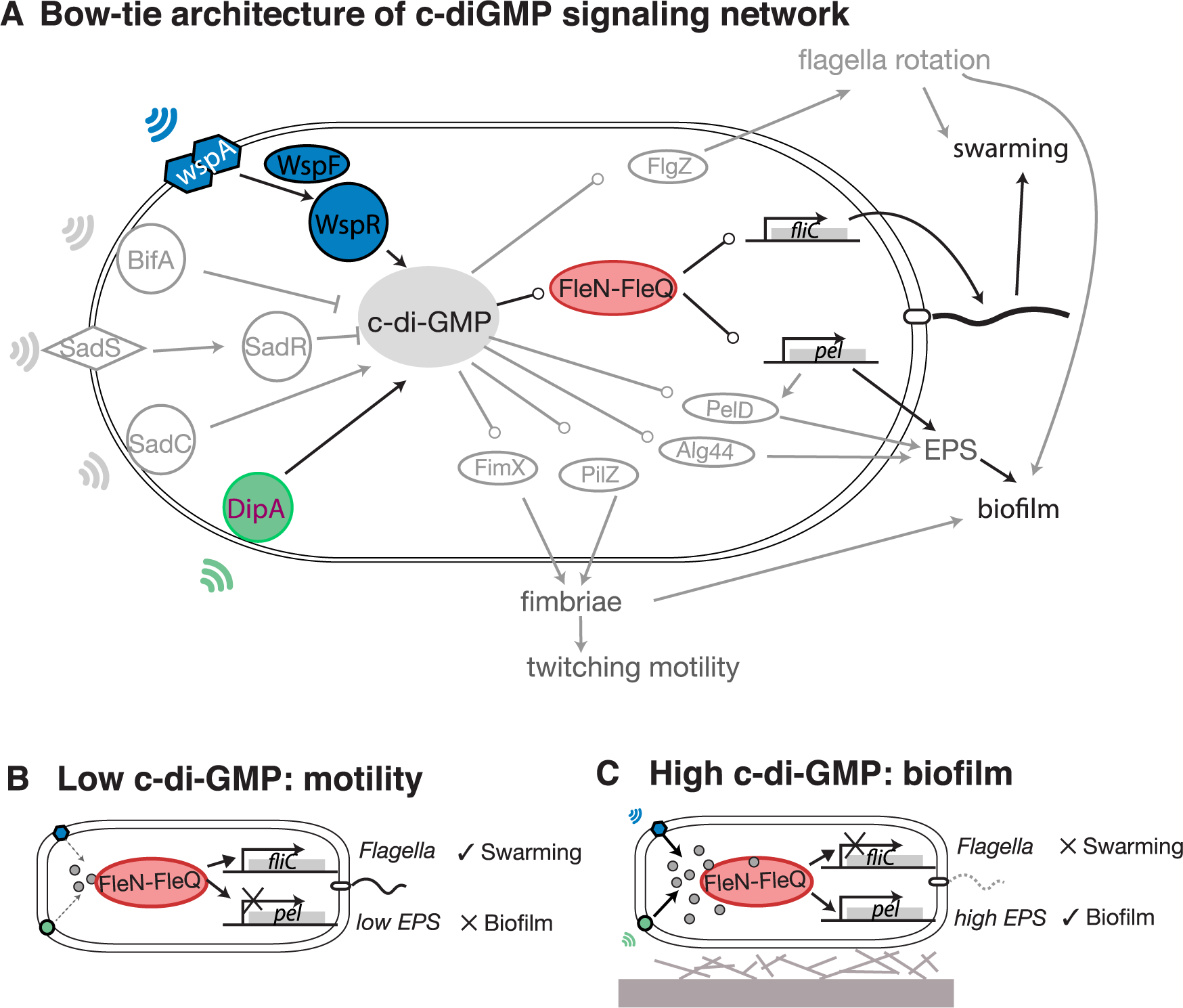
![Hernandez, Bernstein, Pagan, Vargas, McCaig, Ramrattan, Akther, Larracuente, Di, Vieira, Qiu. (2018). BbWrapper: BioPerl-based sequence and tree utilities for rapid prototyping of bioinformatics pipelines. BMC Bioinformatics 19(1):76. [PMID 29499649]](/images/a/a5/12859_2018_2074_Fig1_HTML.png)
![Di, Pagan, Packer, Martin, Akther, Ramrattan, Mongodin, Fraser, Schutzer, Luft, Casjens and Qiu. (2014). BorreliaBase: a phylogeny-centered browser of Borrelia genomes. BMC Bioinformatics 15:233. [PMID 24994456]](/images/8/8d/12859_2014_Article_6488_Fig3_HTML.png)